Leapfrog dynamics in phage-bacteria coevolution revealed by joint analysis of cross-infection phenotypes and whole genome sequencing
- PMID: 35092147
- PMCID: PMC10167754
- DOI: 10.1111/ele.13965
Leapfrog dynamics in phage-bacteria coevolution revealed by joint analysis of cross-infection phenotypes and whole genome sequencing
Abstract
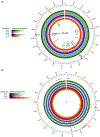
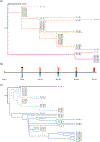
Viruses and their hosts can undergo coevolutionary arms races where hosts evolve increased resistance and viruses evolve counter-resistance. Given these arms race dynamics (ARD), both players are predicted to evolve along a single trajectory as more recently evolved genotypes replace their predecessors. By coupling phenotypic and genomic analyses of coevolving populations of bacteriophage λ and Escherichia coli, we find conflicting evidence for ARD. Virus-host infection phenotypes fit the ARD model, yet genomic analyses revealed fluctuating selection dynamics. Rather than coevolution unfolding along a single trajectory, cryptic genetic variation emerges and is maintained at low frequency for generations until it eventually supplants dominant lineages. These observations suggest a hybrid 'leapfrog' dynamic, revealing weaknesses in the predictive power of standard coevolutionary models. The findings shed light on the mechanisms that structure coevolving ecological networks and reveal the limits of using phenotypic or genomic data alone to differentiate coevolutionary dynamics.
© 2022 John Wiley & Sons Ltd.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no competing interests.
Figures

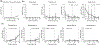
References
-
- Adams MH (1959) Bacteriophages. New York: Interscience Publishers Inc.
-
- Agrawal A & Lively CM (2002) Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evolutionary Ecology Research, 4, 79–90.
-
- Almeida-Neto M, Guimarães P, Guimarães PR Jr, Loyola RD & Ulrich W (2008) A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos, 117, 1227–1239.
-
- Andrews S (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Accessed May 2018].
-
- Ashby B, Iritani R, Best A, White A & Boots M (2019) Understanding the role of eco-evolutionary feedbacks in host-parasite coevolution. Journal of Theoretical Biology, 464, 115–125. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

