Mutations that adapt SARS-CoV-2 to mink or ferret do not increase fitness in the human airway
- PMID: 35093235
- PMCID: PMC8768428
- DOI: 10.1016/j.celrep.2022.110344
Mutations that adapt SARS-CoV-2 to mink or ferret do not increase fitness in the human airway
Abstract
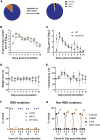
SARS-CoV-2 has a broad mammalian species tropism infecting humans, cats, dogs, and farmed mink. Since the start of the 2019 pandemic, several reverse zoonotic outbreaks of SARS-CoV-2 have occurred in mink, one of which reinfected humans and caused a cluster of infections in Denmark. Here we investigate the molecular basis of mink and ferret adaptation and demonstrate the spike mutations Y453F, F486L, and N501T all specifically adapt SARS-CoV-2 to use mustelid ACE2. Furthermore, we risk assess these mutations and conclude mink-adapted viruses are unlikely to pose an increased threat to humans, as Y453F attenuates the virus replication in human cells and all three mink adaptations have minimal antigenic impact. Finally, we show that certain SARS-CoV-2 variants emerging from circulation in humans may naturally have a greater propensity to infect mustelid hosts and therefore these species should continue to be surveyed for reverse zoonotic infections.
Keywords: ACE2; COVID-19; SARS-CoV-2; antigenicity; coronavirus; ferret; mink; pandemic; zoonosis.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Author Anonymous . 2021. Poland Orders Cull at Fur Farm with Country’s First Mink Coronavirus Case (Notes from Poland)https://notesfrompoland.com/2021/02/01/poland-orders-cull-at-fur-farm-wi...
-
- Author Anonymous . Spanish News Today. 2021. Renewed calls for closure of Galicia mink farms after four more Covid outbreaks.https://spanishnewstoday.com/renewed-calls-for-closure-of-galicia-mink-f...
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- BBS/E/I/00007034/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/W005611/1/MRC_/Medical Research Council/United Kingdom
- BBS/E/I/COV07001/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UU_12014/10/MRC_/Medical Research Council/United Kingdom
- MC_PC_19026/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

