Integrated Liver and Plasma Proteomics in Obese Mice Reveals Complex Metabolic Regulation
- PMID: 35093608
- PMCID: PMC8928073
- DOI: 10.1016/j.mcpro.2022.100207
Integrated Liver and Plasma Proteomics in Obese Mice Reveals Complex Metabolic Regulation
Abstract
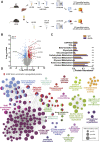
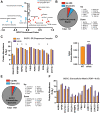
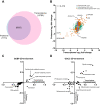
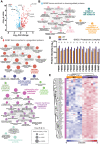
Obesity leads to the development of nonalcoholic fatty liver disease (NAFLD) and associated alterations to the plasma proteome. To elucidate the underlying changes associated with obesity, we performed liquid chromatography-tandem mass spectrometry in the liver and plasma of obese leptin-deficient ob/ob mice and integrated these data with publicly available transcriptomic and proteomic datasets of obesity and metabolic diseases in preclinical and clinical cohorts. We quantified 7173 and 555 proteins in the liver and plasma proteomes, respectively. The abundance of proteins related to fatty acid metabolism were increased, alongside peroxisomal proliferation in ob/ob liver. Putatively secreted proteins and the secretory machinery were also dysregulated in the liver, which was mirrored by a substantial alteration of the plasma proteome. Greater than 50% of the plasma proteins were differentially regulated, including NAFLD biomarkers, lipoproteins, the 20S proteasome, and the complement and coagulation cascades of the immune system. Integration of the liver and plasma proteomes identified proteins that were concomitantly regulated in the liver and plasma in obesity, suggesting that the systemic abundance of these plasma proteins is regulated by secretion from the liver. Many of these proteins are systemically regulated during type 2 diabetes and/or NAFLD in humans, indicating the clinical importance of liver-plasma cross talk and the relevance of our investigations in ob/ob mice. Together, these analyses yield a comprehensive insight into obesity and provide an extensive resource for obesity research in a prevailing model organism.
Keywords: NAFLD; cross talk; leptin; obesity; proteome.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures




References
-
- Gruben N., Shiri-Sverdlov R., Koonen D.P.Y., Hofker M.H. Nonalcoholic fatty liver disease: A main driver of insulin resistance or a dangerous liaison? Biochim. Biophys. Acta. 2014;1842:2329–2343. - PubMed
-
- Knobler H., Schattner A., Zhornicki T., Malnick S.D., Keter D., Sokolovskaya N., Lurie Y., Bass D.D. Fatty liver–an additional and treatable feature of the insulin resistance syndrome. QJM. 1999;92:73–79. - PubMed
-
- Utzschneider K.M., Kahn S.E. The role of insulin resistance in nonalcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 2006;91:4753–4761. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

