The Role of Microtubule Associated Serine/Threonine Kinase 3 Variants in Neurodevelopmental Diseases: Genotype-Phenotype Association
- PMID: 35095415
- PMCID: PMC8790505
- DOI: 10.3389/fnmol.2021.775479
The Role of Microtubule Associated Serine/Threonine Kinase 3 Variants in Neurodevelopmental Diseases: Genotype-Phenotype Association
Abstract
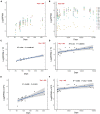
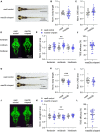
Objective: To prove microtubule associated serine/threonine kinase 3 (MAST3) gene is associated with neurodevelopmental diseases (NDD) and the genotype-phenotype correlation. Methods: Trio exome sequencing (trio ES) was performed on four NDD trios. Bioinformatic analysis was conducted based on large-scale genome sequencing data and human brain transcriptomic data. Further in vivo zebrafish studies were performed. Results: In our study, we identified four de novo MAST3 variants (NM_015016.1: c.302C > T:p.Ser101Phe; c.311C > T:p.Ser104Leu; c.1543G > A:p.Gly515Ser; and c.1547T > C:p.Leu516Pro) in four patients with developmental and epileptic encephalopathy (DEE) separately. Clinical heterogeneities were observed in patients carrying variants in domain of unknown function (DUF) and serine-threonine kinase (STK) domain separately. Using the published large-scale exome sequencing data, higher CADD scores of missense variants in DUF domain were found in NDD cohort compared with gnomAD database. In addition, we obtained an excess of missense variants in DUF domain when compared autistic spectrum disorder (ASD) cohort with gnomAD database, similarly an excess of missense variants in STK domain when compared DEE cohort with gnomAD database. Based on Brainspan datasets, we showed that MAST3 expression was significantly upregulated in ASD and DEE-related brain regions and was functionally linked with DEE genes. In zebrafish model, abnormal morphology of central nervous system was observed in mast3a/b crispants. Conclusion: Our results support the possibility that MAST3 is a novel gene associated with NDD which could expand the genetic spectrum for NDD. The genotype-phenotype correlation may contribute to future genetic counseling.
Keywords: MAST3; domain; epilepsy; genetics; neurodevelopmental.
Copyright © 2022 Shu, Xiao, Qin, Tian, Zhang, Li, Liu, Li, Gu, Wang, Wang and Mao.
Conflict of interest statement
JL was employed by the company Cipher Gene LLC. WG and PW were employed by the company Chigene (Beijing) Translational Medical Research Center Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Brant B., Stern T., Shekhidem H. A., Mizrahi L., Rosh I., Stern Y., et al. (2021). IQSEC2 mutation associated with epilepsy, intellectual disability, and autism results in hyperexcitability of patient-derived neurons and deficient synaptic transmission. Mol. Psychiatry 12:43. 10.1038/s41380-021-01281-0 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

