DNA Double Strand Break Repair and Its Control by Nucleosome Remodeling
- PMID: 35096025
- PMCID: PMC8790285
- DOI: 10.3389/fgene.2021.821543
DNA Double Strand Break Repair and Its Control by Nucleosome Remodeling
Abstract
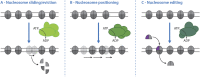
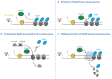
DNA double strand breaks (DSBs) are repaired in eukaryotes by one of several cellular mechanisms. The decision-making process controlling DSB repair takes place at the step of DNA end resection, the nucleolytic processing of DNA ends, which generates single-stranded DNA overhangs. Dependent on the length of the overhang, a corresponding DSB repair mechanism is engaged. Interestingly, nucleosomes-the fundamental unit of chromatin-influence the activity of resection nucleases and nucleosome remodelers have emerged as key regulators of DSB repair. Nucleosome remodelers share a common enzymatic mechanism, but for global genome organization specific remodelers have been shown to exert distinct activities. Specifically, different remodelers have been found to slide and evict, position or edit nucleosomes. It is an open question whether the same remodelers exert the same function also in the context of DSBs. Here, we will review recent advances in our understanding of nucleosome remodelers at DSBs: to what extent nucleosome sliding, eviction, positioning and editing can be observed at DSBs and how these activities affect the DSB repair decision.
Keywords: DNA end resection; DNA repair; cell cycle; double strand break; genome stability; nucleosome remodeling.
Copyright © 2022 Karl, Peritore, Galanti and Pfander.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Adra C. N., Donato J.-L., Badovinac R., Syed F., Kheraj R., Cai H., et al. (2000). SMARCAD1, a Novel Human Helicase Family-Defining Member Associated with Genetic Instability: Cloning, Expression, and Mapping to 4q22-Q23, a Band Rich in Breakpoints and Deletion Mutants Involved in Several Human Diseases. Genomics 69, 162–173. 10.1006/geno.2000.6281 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

