Molecular Analysis of fimA Operon Genes among UPEC Local Isolates in Baghdad City
- PMID: 35096318
- PMCID: PMC8790995
- DOI: 10.22092/ari.2021.355465.1689
Molecular Analysis of fimA Operon Genes among UPEC Local Isolates in Baghdad City
Abstract
Specialized Escherichia coli (E. coli) isolates, called uropathogenic E. coli (UPEC), cause most of urinary tract infections (UITs). Once bacteria reached the urinary tract of the host, they have to adhere to the host cell for the colonization. For this purpose, bacteria have different structures including fimbrial adhesins. Most of the UPECs contain type 1 fimbriae encoded by fim operon (fimB, E, A, I, C, D, F, G, H) which is responsible for the adhesive ability in these isolates. Ninety-four isolates of UPEC were obtained from UTI patients in Baghdad hospitals and their diagnosis were confirmed by the PCR method using 16srDNA as a housekeeping gene. The UPEC isolates were tested for their ability of adherence to the urothelial cells obtained from the mid-stream urine from healthy women. Fifty isolates were subjected to detect type1 fimbriae genes (fimA operon) using specific primers followed by sequencing the amplified fragment which they were analyzed by Geneious software. The results confirm that all the isolates were E. coli according to the genetic analysis by the PCR test, and also, the ability of attachment for all isolates were approved (100%). For type 1 fimbriae, the findings figured out that 100% of the isolates harbored fimA,fimI, fimC, fimD, fimG and fimH genes; while 96% of them were positive for fimB, fimF,and 82% of the isolates were positive for fimE. This result exhibited a higher prevalence of fim genes, as the attachment ability was 100%. Approximately, all UPEC have type 1fimbrial genes, so it could be used as a genetic marker in the investigation of E. coli adhesion ability.
Keywords: Uropathogenic Escherichia coli; attachment ability; fimA operon.
Figures




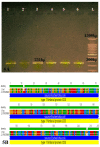





References
-
- Bader MS, Loeb M, Brooks AA. An update on the management of urinary tract infections in the era of antimicrobial resistance. Postgrad Med. 2017;129(2):242–58. - PubMed
-
- Pusz P, Bok E, Mazurek J, Stosik M, Baldy-Chudzik K. Type 1 fimbriae in commensal Escherichia coli derived from healthy humans. Acta Biochim Pol. 2014;61(2) - PubMed
-
- Stærk K, Khandige S, Kolmos HJ, Møller-Jensen J, Andersen TE. Uropathogenic Escherichia coli express type 1 fimbriae only in surface adherent populations under physiological growth conditions. J Infect Dis. 2016;213(3):386–94. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
