Paracrine Behaviors Arbitrate Parasite-Like Interactions Between Tumor Subclones
- PMID: 35096847
- PMCID: PMC8794381
- DOI: 10.3389/fevo.2021.675638
Paracrine Behaviors Arbitrate Parasite-Like Interactions Between Tumor Subclones
Abstract
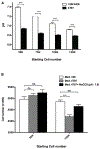
Explaining the emergence and maintenance of intratumor heterogeneity is an important question in cancer biology. Tumor cells can generate considerable subclonal diversity, which influences tumor growth rate, treatment resistance, and metastasis, yet we know remarkably little about how cells from different subclones interact. Here, we confronted two murine mammary cancer cell lines to determine both the nature and mechanisms of subclonal cellular interactions in vitro. Surprisingly, we found that, compared to monoculture, growth of the "winner" was enhanced by the presence of the "loser" cell line, whereas growth of the latter was reduced. Mathematical modeling and laboratory assays indicated that these interactions are mediated by the production of paracrine metabolites resulting in the winner subclone effectively "farming" the loser. Our findings add a new level of complexity to the mechanisms underlying subclonal growth dynamics.
Keywords: Lotka–Volterra model; beta-hydroxybutirate; cancer evolution; evolutionary game theory; intratumor clonal heterogeneity; lactate; paracrine signaling; parasitism.
Conflict of interest statement
Conflict of Interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Amirouchene-Angelozzi N, Swanton C, and Bardelli A (2017). Tumor evolution as a therapeutic target. Cancer Discov. 7, 805–817. doi: 10.1158/2159-8290.cd-17-0343 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
