Novel stress granule-like structures are induced via a paracrine mechanism during viral infection
- PMID: 35098996
- PMCID: PMC8976915
- DOI: 10.1242/jcs.259194
Novel stress granule-like structures are induced via a paracrine mechanism during viral infection
Abstract
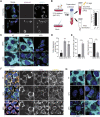
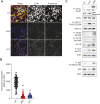
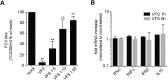
To rapidly adapt to stresses such as infections, cells have evolved several mechanisms, which include the activation of stress response pathways and the innate immune response. These stress responses result in the rapid inhibition of translation and condensation of stalled mRNAs with RNA-binding proteins and signalling components into cytoplasmic biocondensates called stress granules (SGs). Increasing evidence suggests that SGs contribute to antiviral defence, and thus viruses need to evade these responses to propagate. We previously showed that feline calicivirus (FCV) impairs SG assembly by cleaving the scaffolding protein G3BP1. We also observed that uninfected bystander cells assembled G3BP1-positive granules, suggesting a paracrine response triggered by infection. We now present evidence that virus-free supernatant generated from infected cells can induce the formation of SG-like foci, which we name paracrine granules. They are linked to antiviral activity and exhibit specific kinetics of assembly-disassembly, and protein and RNA composition that are different from canonical SGs. We propose that this paracrine induction reflects a novel cellular defence mechanism to limit viral propagation and promote stress responses in bystander cells.
Keywords: G3BP1; Stress granule; Virus.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

