Heinz-resistant tomato cultivars exhibit a lignin-based resistance to field dodder (Cuscuta campestris) parasitism
- PMID: 35099559
- PMCID: PMC9070836
- DOI: 10.1093/plphys/kiac024
Heinz-resistant tomato cultivars exhibit a lignin-based resistance to field dodder (Cuscuta campestris) parasitism
Abstract
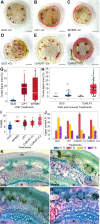
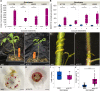
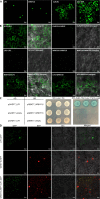
Cuscuta species (dodders) are agriculturally destructive, parasitic angiosperms. These parasitic plants use haustoria as physiological bridges to extract nutrients and water from hosts. Cuscuta campestris has a broad host range and wide geographical distribution. While some wild tomato relatives are resistant, cultivated tomatoes are generally susceptible to C. campestris infestations. However, some specific Heinz tomato (Solanum lycopersicum) hybrid cultivars exhibit resistance to dodders in the field, but their defense mechanism was previously unknown. Here, we discovered that the stem cortex in these resistant lines responds with local lignification upon C. campestris attachment, preventing parasite entry into the host. Lignin Induction Factor 1 (LIF1, an AP2-like transcription factor), SlMYB55, and Cuscuta R-gene for Lignin-based Resistance 1, a CC-NBS-LRR (CuRLR1) are identified as factors that confer host resistance by regulating lignification. SlWRKY16 is upregulated upon C. campestris infestation and potentially negatively regulates LIF1 function. Intriguingly, CuRLR1 may play a role in signaling or function as an intracellular receptor for receiving Cuscuta signals or effectors, thereby regulating lignification-based resistance. In summary, these four regulators control the lignin-based resistance response in specific Heinz tomato cultivars, preventing C. campestris from parasitizing resistant tomatoes. This discovery provides a foundation for investigating multilayer resistance against Cuscuta species and has potential for application in other essential crops attacked by parasitic plants.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures



Similar articles
-
Investigating Host and Parasitic Plant Interaction by Tissue-Specific Gene Analyses on Tomato and Cuscuta campestris Interface at Three Haustorial Developmental Stages.Front Plant Sci. 2022 Feb 10;12:764843. doi: 10.3389/fpls.2021.764843. eCollection 2021. Front Plant Sci. 2022. PMID: 35222447 Free PMC article.
-
Identification of tomato introgression lines with enhanced susceptibility or resistance to infection by parasitic giant dodder (Cuscuta reflexa).Physiol Plant. 2018 Feb;162(2):205-218. doi: 10.1111/ppl.12660. Epub 2017 Dec 8. Physiol Plant. 2018. PMID: 29080211
-
Cuscuta campestris fine-tunes gene expression during haustoriogenesis as an adaptation to different hosts.Plant Physiol. 2023 Dec 30;194(1):258-273. doi: 10.1093/plphys/kiad505. Plant Physiol. 2023. PMID: 37706590 Free PMC article.
-
Parasitic plants of the genus Cuscuta and their interaction with susceptible and resistant host plants.Front Plant Sci. 2015 Feb 4;6:45. doi: 10.3389/fpls.2015.00045. eCollection 2015. Front Plant Sci. 2015. PMID: 25699071 Free PMC article. Review.
-
Cuscuta species: Model organisms for haustorium development in stem holoparasitic plants.Front Plant Sci. 2022 Dec 12;13:1086384. doi: 10.3389/fpls.2022.1086384. eCollection 2022. Front Plant Sci. 2022. PMID: 36578337 Free PMC article. Review.
Cited by
-
Investigating Host and Parasitic Plant Interaction by Tissue-Specific Gene Analyses on Tomato and Cuscuta campestris Interface at Three Haustorial Developmental Stages.Front Plant Sci. 2022 Feb 10;12:764843. doi: 10.3389/fpls.2021.764843. eCollection 2021. Front Plant Sci. 2022. PMID: 35222447 Free PMC article.
-
Striga hermonthica induces lignin deposition at the root tip to facilitate prehaustorium formation and obligate parasitism.Plant Commun. 2025 May 12;6(5):101294. doi: 10.1016/j.xplc.2025.101294. Epub 2025 Mar 6. Plant Commun. 2025. PMID: 40033692 Free PMC article.
-
Parasite-resistant ketchup! Lignin-based resistance to parasitic plants in tomato.Plant Physiol. 2022 May 3;189(1):4-6. doi: 10.1093/plphys/kiac067. Plant Physiol. 2022. PMID: 35188196 Free PMC article. No abstract available.
-
Metabolic changes induced by Cuscuta campestris Yunck in the host species Artemisia campestris subsp. variabilis (Ten.) Greuter as a strategy for successful parasitisation.Planta. 2022 Nov 15;256(6):118. doi: 10.1007/s00425-022-04025-8. Planta. 2022. PMID: 36376619 Free PMC article.
-
Extracellular Self- and Non-Self DNA Involved in Damage Recognition in the Mistletoe Parasitism of Mesquite Trees.Int J Mol Sci. 2023 Dec 29;25(1):457. doi: 10.3390/ijms25010457. Int J Mol Sci. 2023. PMID: 38203628 Free PMC article.
References
-
- Adcock IM, Caramori G (2009) Transcription factors. In Barnes PJ, Drazen JM, Rennard SI, Thomson NC, eds, Asthma and COPD, Ed 2. Academic Press, Oxford, pp 373–380
-
- Agrios GN (2005) Plant diseases caused by parasitic higher plants, invasive climbing plants, and parasitic green algae. In Plant Pathology, Ed 5. Academic Press, San Diego, pp 705–722
-
- Ashton FM (1976) Cuscuta Spp. (Dodder): A Literature Review of Its Biology and Control. Division of Agricultural Sciences, University of California, Issues pp 1872–1890

