ZNF146/OZF and ZNF507 target LINE-1 sequences
- PMID: 35100360
- PMCID: PMC8896011
- DOI: 10.1093/g3journal/jkac002
ZNF146/OZF and ZNF507 target LINE-1 sequences
Abstract
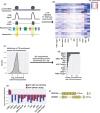
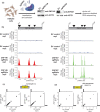
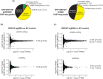
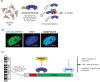
Repetitive sequences including transposable elements and transposon-derived fragments account for nearly half of the human genome. While transposition-competent transposable elements must be repressed to maintain genomic stability, mutated and fragmented transposable elements comprising the bulk of repetitive sequences can also contribute to regulation of host gene expression and broader genome organization. Here, we analyzed published ChIP-seq data sets to identify proteins broadly enriched on transposable elements in the human genome. We show 2 of the proteins identified, C2H2 zinc finger-containing proteins ZNF146 (also known as OZF) and ZNF507, are targeted to distinct sites within LINE-1 ORF2 at thousands of locations in the genome. ZNF146 binding sites are found at old and young LINE-1 elements. In contrast, ZNF507 preferentially binds at young LINE-1 sequences correlated to sequence changes in LINE-1 elements at ZNF507's binding site. To gain further insight into ZNF146 and ZNF507 function, we disrupt their expression in HEK293 cells using CRISPR/Cas9 and perform RNA sequencing, finding modest gene expression changes in cells where ZNF507 has been disrupted. We further identify a physical interaction between ZNF507 and PRMT5, suggesting ZNF507 may target arginine methylation activity to LINE-1 sequences.
Keywords: L1; LINE-1; OZF; PRMT5; ZNF146; ZNF507; transposable element.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures



References
-
- Antoine K, Ferbus D, Kolahgar G, Prosperi MT, Goubin G.. Zinc finger protein overexpressed in colon carcinoma interacts with the telomeric protein hRap1. J Cell Biochem. 2005a;95(4):763–768. - PubMed
-
- Antoine K, Prosperi MT, Ferbus D, Boule C, Goubin G.. A Kruppel zinc finger of ZNF 146 interacts with the SUMO-1 conjugating enzyme UBC9 and is sumoylated in vivo. Mol Cell Biochem. 2005b;271(1–2):215–223. - PubMed
-
- Bailey TL, Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
