spatzie: an R package for identifying significant transcription factor motif co-enrichment from enhancer-promoter interactions
- PMID: 35100401
- PMCID: PMC9122533
- DOI: 10.1093/nar/gkac036
spatzie: an R package for identifying significant transcription factor motif co-enrichment from enhancer-promoter interactions
Abstract
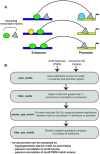
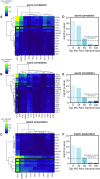
Genomic interactions provide important context to our understanding of the state of the genome. One question is whether specific transcription factor interactions give rise to genome organization. We introduce spatzie, an R package and a website that implements statistical tests for significant transcription factor motif cooperativity between enhancer-promoter interactions. We conducted controlled experiments under realistic simulated data from ChIP-seq to confirm spatzie is capable of discovering co-enriched motif interactions even in noisy conditions. We then use spatzie to investigate cell type specific transcription factor cooperativity within recent human ChIA-PET enhancer-promoter interaction data. The method is available online at https://spatzie.mit.edu.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Kosak S.T., Groudine M.. Form follows function: the genomic organization of cellular differentiation. Genes Dev. 2004; 18:1371–1384. - PubMed
-
- Lanctôt C., Cheutin T., Cremer M., Cavalli G., Cremer T.. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007; 8:104–115. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

