Assembly-dependent translation of subunits 6 (Atp6) and 9 (Atp9) of ATP synthase in yeast mitochondria
- PMID: 35100419
- PMCID: PMC8893259
- DOI: 10.1093/genetics/iyac007
Assembly-dependent translation of subunits 6 (Atp6) and 9 (Atp9) of ATP synthase in yeast mitochondria
Abstract
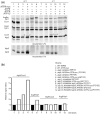
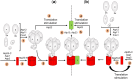
The yeast mitochondrial ATP synthase is an assembly of 28 subunits of 17 types of which 3 (subunits 6, 8, and 9) are encoded by mitochondrial genes, while the 14 others have a nuclear genetic origin. Within the membrane domain (FO) of this enzyme, the subunit 6 and a ring of 10 identical subunits 9 transport protons across the mitochondrial inner membrane coupled to ATP synthesis in the extra-membrane structure (F1) of ATP synthase. As a result of their dual genetic origin, the ATP synthase subunits are synthesized in the cytosol and inside the mitochondrion. How they are produced in the proper stoichiometry from two different cellular compartments is still poorly understood. The experiments herein reported show that the rate of translation of the subunits 9 and 6 is enhanced in strains with mutations leading to specific defects in the assembly of these proteins. These translation modifications involve assembly intermediates interacting with subunits 6 and 9 within the final enzyme and cis-regulatory sequences that control gene expression in the organelle. In addition to enabling a balanced output of the ATP synthase subunits, these assembly-dependent feedback loops are presumably important to limit the accumulation of harmful assembly intermediates that have the potential to dissipate the mitochondrial membrane electrical potential and the main source of chemical energy of the cell.
Keywords: ATP synthase; mitochondria; mitochondria DNA; mitochondrial biogenesis; mitochondrial gene expression; yeast.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Conflict of interest statement
None declared.
Figures






References
-
- Ackerman SH. Atp11p and Atp12p are chaperones for F(1)-ATPase biogenesis in mitochondria. Biochim Biophys Acta. 2002;1555(1–3):101–105. - PubMed
-
- Ackerman SH, Gatti DL, Gellefors P, Douglas MG, Tzagoloff A. ATP13, a nuclear gene of Saccharomyces cerevisiae essential for the expression of subunit 9 of the mitochondrial ATPase. FEBS Lett. 1991;278(2):234–238. - PubMed
-
- Ackerman SH, Tzagoloff A. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem. 1990a;265(17):9952–9959. - PubMed
-
- Ackerman SH, Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog Nucleic Acid Res Mol Biol. 2005;80:95–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

