The regional sequestration of heterochromatin structural proteins is critical to form and maintain silent chromatin
- PMID: 35101096
- PMCID: PMC8805269
- DOI: 10.1186/s13072-022-00435-w
The regional sequestration of heterochromatin structural proteins is critical to form and maintain silent chromatin
Abstract
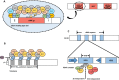
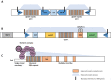
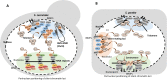
Budding yeast Saccharomyces cerevisiae and fission yeast Schizosaccharomyces pombe are good models for heterochromatin study. In S. pombe, H3K9 methylation and Swi6, an ortholog of mammalian HP1, lead to heterochromatin formation. However, S. cerevisiae does not have known epigenetic silencing markers and instead has Sir proteins to regulate silent chromatin formation. Although S. cerevisiae and S. pombe form and maintain heterochromatin via mechanisms that appear to be fundamentally different, they share important common features in the heterochromatin structural proteins. Heterochromatin loci are localized at the nuclear periphery by binding to perinuclear membrane proteins, thereby producing distinct heterochromatin foci, which sequester heterochromatin structural proteins. In this review, we discuss the nuclear peripheral anchoring of heterochromatin foci and its functional relevance to heterochromatin formation and maintenance.
Keywords: Heterochromatin structural proteins; SIR complex; Saccharomyces cerevisiae; Schizosaccharomyces pombe; Swi6.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Opposing Roles of FACT for Euchromatin and Heterochromatin in Yeast.Biomolecules. 2023 Feb 16;13(2):377. doi: 10.3390/biom13020377. Biomolecules. 2023. PMID: 36830746 Free PMC article. Review.
-
Uncoupling the distinct functions of HP1 proteins during heterochromatin establishment and maintenance.Cell Rep. 2023 Nov 28;42(11):113428. doi: 10.1016/j.celrep.2023.113428. Epub 2023 Nov 11. Cell Rep. 2023. PMID: 37952152 Free PMC article.
-
Characterization of the Swi6/HP1 binding motif in its partner protein reveals the basis for the functional divergence of the HP1 family proteins in fission yeast.FASEB J. 2025 Feb 28;39(4):e70387. doi: 10.1096/fj.202402264RR. FASEB J. 2025. PMID: 39945308 Free PMC article.
-
H3K9 methylation extends across natural boundaries of heterochromatin in the absence of an HP1 protein.EMBO J. 2015 Nov 12;34(22):2789-803. doi: 10.15252/embj.201591320. Epub 2015 Oct 5. EMBO J. 2015. PMID: 26438724 Free PMC article.
-
Destabilizing heterochromatin: Does Swi6/HP1 make the choice?Mol Cell. 2006 Jun 23;22(6):709-710. doi: 10.1016/j.molcel.2006.06.004. Mol Cell. 2006. PMID: 16793539 Review.
Cited by
-
Spt3 and Spt8 Are Involved in the Formation of a Silencing Boundary by Interacting with TATA-Binding Protein.Biomolecules. 2023 Mar 30;13(4):619. doi: 10.3390/biom13040619. Biomolecules. 2023. PMID: 37189367 Free PMC article.
-
Telomere-to-Telomere genome assemblies of human-infecting Encephalitozoon species.BMC Genomics. 2023 May 4;24(1):237. doi: 10.1186/s12864-023-09331-3. BMC Genomics. 2023. PMID: 37142951 Free PMC article.
-
A dual, catalytic role for the fission yeast Ccr4-Not complex in gene silencing and heterochromatin spreading.Genetics. 2023 Aug 9;224(4):iyad108. doi: 10.1093/genetics/iyad108. Genetics. 2023. PMID: 37279920 Free PMC article.
-
Functions of HP1 proteins in transcriptional regulation.Epigenetics Chromatin. 2022 May 7;15(1):14. doi: 10.1186/s13072-022-00453-8. Epigenetics Chromatin. 2022. PMID: 35526078 Free PMC article. Review.
-
Opposing Roles of FACT for Euchromatin and Heterochromatin in Yeast.Biomolecules. 2023 Feb 16;13(2):377. doi: 10.3390/biom13020377. Biomolecules. 2023. PMID: 36830746 Free PMC article. Review.
References
-
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. - PubMed
-
- Huisinga KL, et al. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115:110–122. - PubMed
-
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. - PubMed
-
- Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Lachner M, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

