Rapid 3D bioprinting of a multicellular model recapitulating pterygium microenvironment
- PMID: 35101743
- PMCID: PMC10162446
- DOI: 10.1016/j.biomaterials.2022.121391
Rapid 3D bioprinting of a multicellular model recapitulating pterygium microenvironment
Abstract
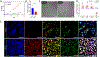
Pterygium is an ocular surface disorder with high prevalence that can lead to vision impairment. As a pathological outgrowth of conjunctiva, pterygium involves neovascularization and chronic inflammation. Here, we developed a 3D multicellular in vitro pterygium model using a digital light processing (DLP)-based 3D bioprinting platform with human conjunctival stem cells (hCjSCs). A novel feeder-free culture system was adopted and efficiently expanded the primary hCjSCs with homogeneity, stemness and differentiation potency. The DLP-based 3D bioprinting method was able to fabricate hydrogel scaffolds that support the viability and biological integrity of the encapsulated hCjSCs. The bioprinted 3D pterygium model consisted of hCjSCs, immune cells, and vascular cells to recapitulate the disease microenvironment. Transcriptomic analysis using RNA sequencing (RNA-seq) identified a distinct profile correlated to inflammation response, angiogenesis, and epithelial mesenchymal transition in the bioprinted 3D pterygium model. In addition, the pterygium signatures and disease relevance of the bioprinted model were validated with the public RNA-seq data from patient-derived pterygium tissues. By integrating the stem cell technology with 3D bioprinting, this is the first reported 3D in vitro disease model for pterygium that can be utilized for future studies towards personalized medicine and drug screening.
Keywords: 3D bioprinting; Disease model; Epithelial mesenchymal transition; Hydrogels; Pterygium; Stem cells; Tissue engineering.
Copyright © 2022 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: SC is a co-founder of and has an equity interest in Allegro 3D, Inc., and he serves on the scientific advisory board. Some of his research grants, including those acknowledged here, have been identified for conflict-of-interest management based on the overall scope of the project and its potential benefit to Allegro 3D, Inc. The author is required to disclose this relationship in publications acknowledging the grant support, however the research subject and findings reported here did not involve the company in any way and have no relationship with the business activities or scientific interests of the company. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict-of-interest policies. The other authors have no competing interests to declare.
Figures
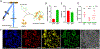

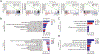
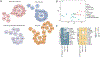
References
-
- Harvey TM, Fernandez AGA, Patel R, Goldman D, Ciralsky J, Conjunctival anatomy and physiology, in: Ocul. Surf. Dis. Cornea, Conjunctiva Tear Film, Elsevier Inc., 2013, pp. 23–27, 10.1016/B978-1-4557-2876-3.00004-3. - DOI
-
- Foster JB, Lee WB, The tear film: anatomy, structure and function, in: Ocul. Surf. Dis. Cornea, Conjunctiva Tear Film, Elsevier Inc., 2013, pp. 17–21, 10.1016/B978-1-4557-2876-3.00003-1. - DOI
-
- Thorel D, Ingen-Housz-Oro S, Royer G, Delcampe A, Bellon N, Bodemer C, Welfringer-Morin A, Bremond-Gignac D, Robert MP, Tauber M, Malecaze F, Dereure O, Daien V, Colin A, Bernier C, Couret C, Vabres B, Tetart F, Milpied B, Cornut T, Ben Said B, Burillon C, Cordel N, Beral L, De Prost N, Wolkenstein P, Muraine M, Gueudry J, Management of ocular involvement in the acute phase of Stevens-Johnson syndrome and toxic epidermal necrolysis: French national audit of practices, literature review, and consensus agreement, Orphanet J. Rare Dis 15 (2020) 259, 10.1186/s13023-020-01538-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

