Group 3 innate lymphoid cells produce the growth factor HB-EGF to protect the intestine from TNF-mediated inflammation
- PMID: 35102343
- PMCID: PMC8842850
- DOI: 10.1038/s41590-021-01110-0
Group 3 innate lymphoid cells produce the growth factor HB-EGF to protect the intestine from TNF-mediated inflammation
Abstract
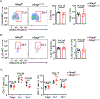
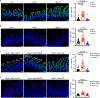
Tumor necrosis factor (TNF) drives chronic inflammation and cell death in the intestine, and blocking TNF is a therapeutic approach in inflammatory bowel disease (IBD). Despite this knowledge, the pathways that protect the intestine from TNF are incompletely understood. Here we demonstrate that group 3 innate lymphoid cells (ILC3s) protect the intestinal epithelium from TNF-induced cell death. This occurs independent of interleukin-22 (IL-22), and we identify that ILC3s are a dominant source of heparin-binding epidermal growth factor-like growth factor (HB-EGF). ILC3s produce HB-EGF in response to prostaglandin E2 (PGE2) and engagement of the EP2 receptor. Mice lacking ILC3-derived HB-EGF exhibit increased susceptibility to TNF-mediated epithelial cell death and experimental intestinal inflammation. Finally, human ILC3s produce HB-EGF and are reduced from the inflamed intestine. These results define an essential role for ILC3-derived HB-EGF in protecting the intestine from TNF and indicate that disruption of this pathway contributes to IBD.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures






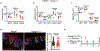







Comment in
-
Intestinal barrier protection.Nat Rev Immunol. 2022 Mar;22(3):144-145. doi: 10.1038/s41577-022-00685-5. Nat Rev Immunol. 2022. PMID: 35105977 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

