Intrinsic entropy model for feature selection of scRNA-seq data
- PMID: 35102420
- PMCID: PMC9175189
- DOI: 10.1093/jmcb/mjac008
Intrinsic entropy model for feature selection of scRNA-seq data
Abstract
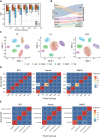
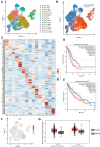
Recent advances of single-cell RNA sequencing (scRNA-seq) technologies have led to extensive study of cellular heterogeneity and cell-to-cell variation. However, the high frequency of dropout events and noise in scRNA-seq data confounds the accuracy of the downstream analysis, i.e. clustering analysis, whose accuracy depends heavily on the selected feature genes. Here, by deriving an entropy decomposition formula, we propose a feature selection method, i.e. an intrinsic entropy (IE) model, to identify the informative genes for accurately clustering analysis. Specifically, by eliminating the 'noisy' fluctuation or extrinsic entropy (EE), we extract the IE of each gene from the total entropy (TE), i.e. TE = IE + EE. We show that the IE of each gene actually reflects the regulatory fluctuation of this gene in a cellular process, and thus high-IE genes provide rich information on cell type or state analysis. To validate the performance of the high-IE genes, we conduct computational analysis on both simulated datasets and real single-cell datasets by comparing with other representative methods. The results show that our IE model is not only broadly applicable and robust for different clustering and classification methods, but also sensitive for novel cell types. Our results also demonstrate that the intrinsic entropy/fluctuation of a gene serves as information rather than noise in contrast to its total entropy/fluctuation.
Keywords: entropy decomposition; extrinsic entropy; feature selection; informative genes; intrinsic entropy; scRNA-seq.
© The Author(s) (2022). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, CEMCS, CAS.
Figures


References
-
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32.
-
- Chen T.Q., Guestrin C. (2016). ‘XGBoost: a scalable tree boosting system’. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, 2016. 785–794. New York, NY, USA: Association for Computing Machinery.
-
- Chen W., Qin Y., Liu S. (2020). CCL20 signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 1231, 53–65. - PubMed

