Dissemination Routes of Carbapenem and Pan-Aminoglycoside Resistance Mechanisms in Hospital and Urban Wastewater Canalizations of Ghana
- PMID: 35103490
- PMCID: PMC8805638
- DOI: 10.1128/msystems.01019-21
Dissemination Routes of Carbapenem and Pan-Aminoglycoside Resistance Mechanisms in Hospital and Urban Wastewater Canalizations of Ghana
Abstract
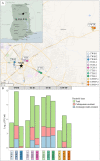
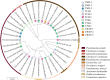
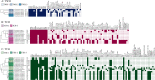
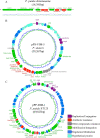
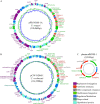
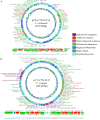
Wastewater has a major role in antimicrobial resistance (AMR) dynamics and public health. The impact on AMR of wastewater flux at the community-hospital interface in low- and middle-income countries (LMICs) is poorly understood. Therefore, the present study analyzed the epidemiological scenario of resistance genes, mobile genetic elements (MGEs), and bacterial populations in wastewater around the Tamale metropolitan area (Ghana). Wastewater samples were collected from the drainage and canalizations before and after three hospitals and one urban waste treatment plant (UWTP). From all carbapenem/pan-aminoglycoside-resistant bacteria, 36 isolates were selected to determine bacterial species and phenotypical resistance profiles. Nanopore sequencing was used to screen resistance genes and plasmids, whereas, sequence types, resistome and plasmidome contents, pan-genome structures, and resistance gene variants were analyzed with Illumina sequencing. The combination of these sequencing data allowed for the resolution of the resistance gene-carrying platforms. Hospitals and the UWTP collected genetic and bacterial elements from community wastewater and amplified successful resistance gene-bacterium associations, which reached the community canalizations. Uncommon carbapenemase/β-lactamase gene variants, like blaDIM-1, and novel variants, including blaVIM-71, blaCARB-53, and blaCMY-172, were identified and seem to spread via clonal expansion of environmental Pseudomonas spp. However, blaNDM-1, blaCTX-M-15, and armA genes, among others, were associated with MGEs that allowed for their dissemination between environmental and clinical bacterial hosts. In conclusion, untreated hospital wastewater in Ghana is a hot spot for the emergence and spread of genes and gene-plasmid-bacterium associations that accelerate AMR, including to last-resort antibiotics. Urgent actions must be taken in wastewater management in LMICs in order to delay AMR expansion. IMPORTANCE Antimicrobial resistance (AMR) is one the major threats to public health today, especially resistance to last-resort compounds for the treatment of critical infections, such as carbapenems and aminoglycosides. Innumerable works have focused on the clinical ambit of AMR, but studies addressing the impact of wastewater cycles on the emergence and dissemination of resistant bacteria are still limited. The lack of knowledge is even greater when referring to low- and middle-income countries, where there is an absence of accurate sanitary systems. Furthermore, the combination of short- and long-read sequencing has surpassed former technical limitations, allowing the complete characterization of resistance genes, mobile genetic platforms, plasmids, and bacteria. The present study deciphered the multiple elements and routes involved in AMR dynamics in wastewater canalizations and, therefore, in the local population of Tamale, providing the basis to adopt accurate control measures to preserve and promote public health.
Keywords: antibiotic resistance; environmental microbiology; genomic analyses; plasmid-mediated resistance; public health; wastewater treatment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Maina M, Tosas-Auguet O, McKnight J, Zosi M, Kimemia G, Mwaniki P, Schultsz C, English M. 2019. Evaluating the foundations that help avert antimicrobial resistance: performance of essential water sanitation and hygiene functions in hospitals and requirements for action in Kenya. PLoS One 14:e0222922. doi:10.1371/journal.pone.0222922. - DOI - PMC - PubMed
-
- Zurfluh K, Bagutti C, Brodmann P, Alt M, Schulze J, Fanning S, Stephan R, Nüesch-Inderbinen M. 2017. Wastewater is a reservoir for clinically relevant carbapenemase- and 16S rRNA methylase-producing Enterobacteriaceae. Int J Antimicrob Agents 50:436–440. doi:10.1016/j.ijantimicag.2017.04.017. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
