Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant
- PMID: 35104835
- PMCID: PMC8942852
- DOI: 10.1038/s41586-022-04462-1
Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant
Abstract
The emergence of the Omicron variant of SARS-CoV-2 is an urgent global health concern1. In this study, our statistical modelling suggests that Omicron has spread more rapidly than the Delta variant in several countries including South Africa. Cell culture experiments showed Omicron to be less fusogenic than Delta and than an ancestral strain of SARS-CoV-2. Although the spike (S) protein of Delta is efficiently cleaved into two subunits, which facilitates cell-cell fusion2,3, the Omicron S protein was less efficiently cleaved compared to the S proteins of Delta and ancestral SARS-CoV-2. Furthermore, in a hamster model, Omicron showed decreased lung infectivity and was less pathogenic compared to Delta and ancestral SARS-CoV-2. Our multiscale investigations reveal the virological characteristics of Omicron, including rapid growth in the human population, lower fusogenicity and attenuated pathogenicity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
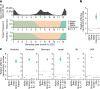
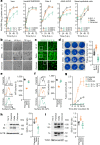


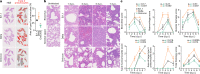

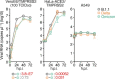





Comment in
-
Omicron: increased transmissibility and decreased pathogenicity.Signal Transduct Target Ther. 2022 May 7;7(1):151. doi: 10.1038/s41392-022-01009-8. Signal Transduct Target Ther. 2022. PMID: 35525870 Free PMC article. No abstract available.
References
-
- WHO. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern; https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.... (2021).
-
- WHO. Tracking SARS-CoV-2 variants; https://www.who.int/en/activities/tracking-SARS-CoV-2-variants (2021).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

