Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity
- PMID: 35104837
- PMCID: PMC8942856
- DOI: 10.1038/s41586-022-04474-x
Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity
Abstract
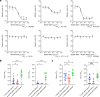
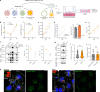
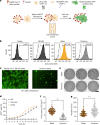
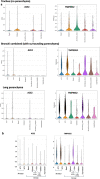
The SARS-CoV-2 Omicron BA.1 variant emerged in 20211 and has multiple mutations in its spike protein2. Here we show that the spike protein of Omicron has a higher affinity for ACE2 compared with Delta, and a marked change in its antigenicity increases Omicron's evasion of therapeutic monoclonal and vaccine-elicited polyclonal neutralizing antibodies after two doses. mRNA vaccination as a third vaccine dose rescues and broadens neutralization. Importantly, the antiviral drugs remdesivir and molnupiravir retain efficacy against Omicron BA.1. Replication was similar for Omicron and Delta virus isolates in human nasal epithelial cultures. However, in lung cells and gut cells, Omicron demonstrated lower replication. Omicron spike protein was less efficiently cleaved compared with Delta. The differences in replication were mapped to the entry efficiency of the virus on the basis of spike-pseudotyped virus assays. The defect in entry of Omicron pseudotyped virus to specific cell types effectively correlated with higher cellular RNA expression of TMPRSS2, and deletion of TMPRSS2 affected Delta entry to a greater extent than Omicron. Furthermore, drug inhibitors targeting specific entry pathways3 demonstrated that the Omicron spike inefficiently uses the cellular protease TMPRSS2, which promotes cell entry through plasma membrane fusion, with greater dependency on cell entry through the endocytic pathway. Consistent with suboptimal S1/S2 cleavage and inability to use TMPRSS2, syncytium formation by the Omicron spike was substantially impaired compared with the Delta spike. The less efficient spike cleavage of Omicron at S1/S2 is associated with a shift in cellular tropism away from TMPRSS2-expressing cells, with implications for altered pathogenesis.
© 2022. The Author(s).
Conflict of interest statement
M.S.P., D.C. and A.d.M. are employees of Vir Biotechnology and may hold shares in Vir Biotechnology. R.K.G. has received honoraria for educational activities from Johnson & Johnson, ViiV and GSK. F.S. is shareholder in Bilitech. The Veesler laboratory has received an unrelated research sponsored agreement from Vir Biotechnology. KGCS is a member of the GSK Immunology Scientific Advisory Board, is a founder, Chief Medical Officer, and a non-executive director of PredictImmune, and is a co-founder of Rheos Medicines. The other authors declare no competing interests.
Figures








Update of
-
SARS-CoV-2 Omicron spike mediated immune escape and tropism shift.Res Sq [Preprint]. 2022 Jan 17:rs.3.rs-1191837. doi: 10.21203/rs.3.rs-1191837/v1. Res Sq. 2022. Update in: Nature. 2022 Mar;603(7902):706-714. doi: 10.1038/s41586-022-04474-x. PMID: 35075452 Free PMC article. Updated. Preprint.
Comment in
-
Omicron: increased transmissibility and decreased pathogenicity.Signal Transduct Target Ther. 2022 May 7;7(1):151. doi: 10.1038/s41392-022-01009-8. Signal Transduct Target Ther. 2022. PMID: 35525870 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- MC_PC_17230/MRC_/Medical Research Council/United Kingdom
- DP1 AI158186/AI/NIAID NIH HHS/United States
- MR/W005611/1/MRC_/Medical Research Council/United Kingdom
- 200594/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI138546/AI/NIAID NIH HHS/United States
- MC_U105181010/MRC_/Medical Research Council/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- MR/T032413/1/MRC_/Medical Research Council/United Kingdom
- MC_UP_1201/16/MRC_/Medical Research Council/United Kingdom
- HHSN272201700059C/AI/NIAID NIH HHS/United States
- WT108082AIA/WT_/Wellcome Trust/United Kingdom
- T32 GM008268/GM/NIGMS NIH HHS/United States
- 207498/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MR/P008801/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

