MUON: multimodal omics analysis framework
- PMID: 35105358
- PMCID: PMC8805324
- DOI: 10.1186/s13059-021-02577-8
MUON: multimodal omics analysis framework
Abstract
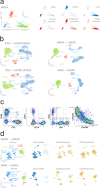
Advances in multi-omics have led to an explosion of multimodal datasets to address questions from basic biology to translation. While these data provide novel opportunities for discovery, they also pose management and analysis challenges, thus motivating the development of tailored computational solutions. Here, we present a data standard and an analysis framework for multi-omics, MUON, designed to organise, analyse, visualise, and exchange multimodal data. MUON stores multimodal data in an efficient yet flexible and interoperable data structure. MUON enables a versatile range of analyses, from data preprocessing to flexible multi-omics alignment.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Argelaguet R, Cuomo ASE, Stegle O. Marioni JC. Computational principles and challenges in single-cell data integration. Nat Biotechnol. 2021. 10.1038/s41587-021-00895-7. - PubMed
-
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3(1):160018. doi: 10.1038/sdata.2016.18. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

