Response and resilience of anammox consortia to nutrient starvation
- PMID: 35105385
- PMCID: PMC8805231
- DOI: 10.1186/s40168-021-01212-9
Response and resilience of anammox consortia to nutrient starvation
Abstract
Background: It is of critical importance to understand how anammox consortia respond to disturbance events and fluctuations in the wastewater treatment reactors. Although the responses of anammox consortia to operational parameters (e.g., temperature, dissolved oxygen, nutrient concentrations) have frequently been reported in previous studies, less is known about their responses and resilience when they suffer from nutrient interruption.
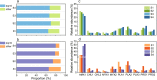
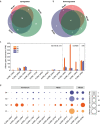
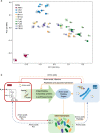
Results: Here, we investigated the anammox community states and transcriptional patterns before and after a short-term nutrient starvation (3 days) to determine how anammox consortia respond to and recover from such stress. The results demonstrated that the remarkable changes in transcriptional patterns, rather than the community compositions were associated with the nutritional stress. The divergent expression of genes involved in anammox reactions, especially the hydrazine synthase complex (HZS), and nutrient transportation might function as part of a starvation response mechanism in anammox bacteria. In addition, effective energy conservation and substrate supply strategies (ATP accumulation, upregulated amino acid biosynthesis, and enhanced protein degradation) and synergistic interactions between anammox bacteria and heterotrophs might benefit their survival during starvation and the ensuing recovery of the anammox process. Compared with abundant heterotrophs in the anammox system, the overall transcription pattern of the core autotrophic producers (i.e., anammox bacteria) was highly resilient and quickly returned to its pre-starvation state, further contributing to the prompt recovery when the feeding was resumed.
Conclusions: These findings provide important insights into nutritional stress-induced changes in transcriptional activities in the anammox consortia and would be beneficial for the understanding of the capacity of anammox consortia in response to stress and process stability in the engineered ecosystems. Video Abstract.
Keywords: Anammox consortia; Nutrient starvation; Recovery; Resilience; Response; Transcriptional pattern.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Lackner S, Gilbert EM, Vlaeminck SE, Joss A, Horn H, van Loosdrecht MCM. Full-scale partial nitritation/anammox experiences—an application survey. Water Res. 2014;55:292–303. - PubMed
-
- Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, et al. Molecular mechanism of anaerobic ammonium oxidation. Nature. 2011;479:127–130. - PubMed
-
- Jin R-C, Yang G-f. Yu J, Zheng P. the inhibition of the Anammox process: a review. Chem Eng J. 2012;197:67–79.
-
- Kuenen JG. Anammox bacteria: from discovery to application. Nat Rev Microbiol. 2008;6:320–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

