Generation of the organotypic kidney structure by integrating pluripotent stem cell-derived renal stroma
- PMID: 35105870
- PMCID: PMC8807595
- DOI: 10.1038/s41467-022-28226-7
Generation of the organotypic kidney structure by integrating pluripotent stem cell-derived renal stroma
Erratum in
-
Author Correction: Generation of the organotypic kidney structure by integrating pluripotent stem cell-derived renal stroma.Nat Commun. 2023 Apr 4;14(1):1874. doi: 10.1038/s41467-023-37484-y. Nat Commun. 2023. PMID: 37015999 Free PMC article. No abstract available.
Abstract
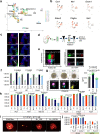
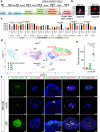
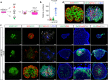
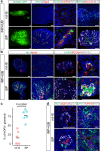
Organs consist of the parenchyma and stroma, the latter of which coordinates the generation of organotypic structures. Despite recent advances in organoid technology, induction of organ-specific stroma and recapitulation of complex organ configurations from pluripotent stem cells (PSCs) have remained challenging. By elucidating the in vivo molecular features of the renal stromal lineage at a single-cell resolution level, we herein establish an in vitro induction protocol for stromal progenitors (SPs) from mouse PSCs. When the induced SPs are assembled with two differentially induced parenchymal progenitors (nephron progenitors and ureteric buds), the completely PSC-derived organoids reproduce the complex kidney structure, with multiple types of stromal cells distributed along differentiating nephrons and branching ureteric buds. Thus, integration of PSC-derived lineage-specific stroma into parenchymal organoids will pave the way toward recapitulation of the organotypic architecture and functions.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

