SARS-CoV-2 genomes from Saudi Arabia implicate nucleocapsid mutations in host response and increased viral load
- PMID: 35105893
- PMCID: PMC8807822
- DOI: 10.1038/s41467-022-28287-8
SARS-CoV-2 genomes from Saudi Arabia implicate nucleocapsid mutations in host response and increased viral load
Abstract
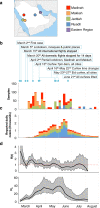
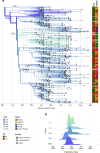
Monitoring SARS-CoV-2 spread and evolution through genome sequencing is essential in handling the COVID-19 pandemic. Here, we sequenced 892 SARS-CoV-2 genomes collected from patients in Saudi Arabia from March to August 2020. We show that two consecutive mutations (R203K/G204R) in the nucleocapsid (N) protein are associated with higher viral loads in COVID-19 patients. Our comparative biochemical analysis reveals that the mutant N protein displays enhanced viral RNA binding and differential interaction with key host proteins. We found increased interaction of GSK3A kinase simultaneously with hyper-phosphorylation of the adjacent serine site (S206) in the mutant N protein. Furthermore, the host cell transcriptome analysis suggests that the mutant N protein produces dysregulated interferon response genes. Here, we provide crucial information in linking the R203K/G204R mutations in the N protein to modulations of host-virus interactions and underline the potential of the nucleocapsid protein as a drug target during infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Organization, W. H. Coronavirus Disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Updatewww.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (2020).
-
- Center, J. H. U. M. C. R. COVID-19 Dashboardhttps://coronavirus.jhu.edu/map.html (2020).
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_19012/MRC_/Medical Research Council/United Kingdom
- Rapid Research Response Team (R3T) by Vice President - Research (VPR) office in KAUST./King Abdullah University of Science and Technology (KAUST)
- MR/R015600/1/MRC_/Medical Research Council/United Kingdom
- KAUST faculty baseline fund (BAS/1/1020-01-01)/King Abdullah University of Science and Technology (KAUST)
- Proposal number: 5-20-01-002-0008/King Abdulaziz City for Science and Technology (KACST)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

