HIV reservoir quantification using cross-subtype multiplex ddPCR
- PMID: 35106463
- PMCID: PMC8786636
- DOI: 10.1016/j.isci.2021.103615
HIV reservoir quantification using cross-subtype multiplex ddPCR
Abstract
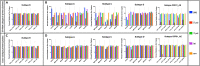
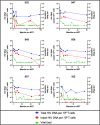
A major barrier to conducting HIV cure research in populations with the highest HIV burden is the lack of an accurate assay to quantify the replication-competent reservoir across the dominant global HIV-1 subtypes. Here, we modify a subtype B HIV-1 assay that quantifies both intact and defective proviral DNA, adapting it to accommodate cross-subtype HIV-1 sequence diversity. We show that the cross-subtype assay works on subtypes A, B, C, D, and CRF01_AE and can detect a single copy of intact provirus. In longitudinal blood samples from Kenyan infants infected with subtypes A and D, patterns of intact and total HIV DNA follow the decay of plasma viral load over time during antiretroviral therapy, with intact HIV DNA comprising 7% (range 1%-33%) of the total HIV DNA during HIV RNA suppression. This high-throughput cross-subtype reservoir assay will be useful in HIV cure research in Africa and Asia, where HIV prevalence is highest.
Keywords: Genotyping; Virology.
© 2021 The Author(s).
Figures


References
-
- Benki S., McClelland R.S., Emery S., Baeten J.M., Richardson B.A., Lavreys L., Mandaliya K., Overbaugh J. Quantification of genital human immunodeficiency virus type 1 (HIV-1) DNA in specimens from women with low plasma HIV-1 RNA levels typical of HIV-1 nontransmitters. J. Clin. Microbiol. 2006;44:4357–4362. - PMC - PubMed
-
- Blankson J.N., Finzi D., Pierson T.C., Sabundayo B.P., Chadwick K., Margolick J.B., Quinn T.C., Siliciano R.F. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J. Infect. Dis. 2000;182:1636–1642. - PubMed
-
- Brown B.K., Darden J.M., Tovanabutra S., Oblander T., Frost J., Sanders-Buell E., de Souza M.S., Birx D.L., McCutchan F.E., Polonis V.R. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J. Virol. 2005;79:6089–6101. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

