Authors' attitude toward adopting a new workflow to improve the computability of phenotype publications
- PMID: 35106535
- PMCID: PMC9278328
- DOI: 10.1093/database/baac001
Authors' attitude toward adopting a new workflow to improve the computability of phenotype publications
Abstract
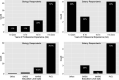
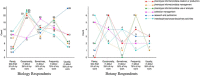
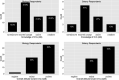
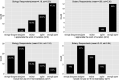
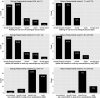
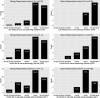
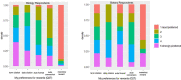
Critical to answering large-scale questions in biology is the integration of knowledge from different disciplines into a coherent, computable whole. Controlled vocabularies such as ontologies represent a clear path toward this goal. Using survey questionnaires, we examined the attitudes of biologists toward adopting controlled vocabularies in phenotype publications. Our questions cover current experience and overall attitude with controlled vocabularies, the awareness of the issues around ambiguity and inconsistency in phenotype descriptions and post-publication professional data curation, the preferred solutions and the effort and desired rewards for adopting a new authoring workflow. Results suggest that although the existence of controlled vocabularies is widespread, their use is not common. A majority of respondents (74%) are frustrated with ambiguity in phenotypic descriptions, and there is a strong agreement (mean agreement score 4.21 out of 5) that author curation would better reflect the original meaning of phenotype data. Moreover, the vast majority (85%) of researchers would try a new authoring workflow if resultant data were more consistent and less ambiguous. Even more respondents (93%) suggested that they would try and possibly adopt a new authoring workflow if it required 5% additional effort as compared to normal, but higher rates resulted in a steep decline in likely adoption rates. Among the four different types of rewards, two types of citations were the most desired incentives for authors to produce computable data. Overall, our results suggest the adoption of a new authoring workflow would be accelerated by a user-friendly and efficient software-authoring tool, an increased awareness of the challenges text ambiguity creates for external curators and an elevated appreciation of the benefits of controlled vocabularies.
© The Author(s) 2022. Published by Oxford University Press.
Figures











References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

