Of mice and human-specific long noncoding RNAs
- PMID: 35106622
- PMCID: PMC8806012
- DOI: 10.1007/s00335-022-09943-2
Of mice and human-specific long noncoding RNAs
Abstract
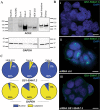
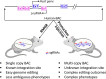
The number of human LncRNAs has now exceeded all known protein-coding genes. Most studies of human LncRNAs have been conducted in cell culture systems where various mechanisms of action have been worked out. On the other hand, efforts to elucidate the function of human LncRNAs in an in vivo setting have been limited. In this brief review, we highlight some strengths and weaknesses of studying human LncRNAs in the mouse. Special consideration is given to bacterial artificial chromosome transgenesis and genome editing. The integration of these technical innovations offers an unprecedented opportunity to complement and extend the expansive literature of cell culture models for the study of human LncRNAs. Two different examples of how BAC transgenesis and genome editing can be leveraged to gain insight into human LncRNA regulation and function in mice are presented: the random integration of a vascular cell-enriched LncRNA and a targeted approach for a new LncRNA immediately upstream of the ACE2 gene, which encodes the receptor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent underlying the coronavirus disease-19 (COVID-19) pandemic.
© 2022. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures



References
-
- Allou L, Balzano S, Magg A, Quinodoz M, Royer-Bertrand B, Schopflin R, et al. Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator. Nature. 2021;592:93–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

