Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome
- PMID: 35108372
- PMCID: PMC11022827
- DOI: 10.1182/blood.2021014472
Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome
Abstract
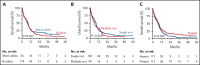
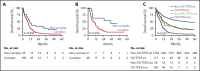
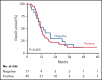
Substantial heterogeneity within mutant TP53 acute myeloid leukemia (AML) and myelodysplastic syndrome with excess of blast (MDS-EB) precludes the exact assessment of prognostic impact for individual patients. We performed in-depth clinical and molecular analysis of mutant TP53 AML and MDS-EB to dissect the molecular characteristics in detail and determine its impact on survival. We performed next-generation sequencing on 2200 AML/MDS-EB specimens and assessed the TP53 mutant allelic status (mono- or bi-allelic), the number of TP53 mutations, mutant TP53 clone size, concurrent mutations, cytogenetics, and mutant TP53 molecular minimal residual disease and studied the associations of these characteristics with overall survival. TP53 mutations were detected in 230 (10.5%) patients with AML/MDS-EB with a median variant allele frequency of 47%. Bi-allelic mutant TP53 status was observed in 174 (76%) patients. Multiple TP53 mutations were found in 49 (21%) patients. Concurrent mutations were detected in 113 (49%) patients. No significant difference in any of the aforementioned molecular characteristics of mutant TP53 was detected between AML and MDS-EB. Patients with mutant TP53 have a poor outcome (2-year overall survival, 12.8%); however, no survival difference between AML and MDS-EB was observed. Importantly, none of the molecular characteristics were significantly associated with survival in mutant TP53 AML/MDS-EB. In most patients, TP53 mutations remained detectable in complete remission by deep sequencing (73%). Detection of residual mutant TP53 was not associated with survival. Mutant TP53 AML and MDS-EB do not differ with respect to molecular characteristics and survival. Therefore, mutant TP53 AML/MDS-EB should be considered a distinct molecular disease entity.
© 2022 by The American Society of Hematology.
Figures

Comment in
-
TP53 and the star-crossed lovers MDS and AML.Blood. 2022 Apr 14;139(15):2265-2266. doi: 10.1182/blood.2022015709. Blood. 2022. PMID: 35420690 Free PMC article. No abstract available.
References
-
- Metzeler KH, Herold T, Rothenberg-Thurley M, et al. AMLCG Study Group Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128(5):686–698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

