Fat3 acts through independent cytoskeletal effectors to coordinate asymmetric cell behaviors during polarized circuit assembly
- PMID: 35108541
- PMCID: PMC8865054
- DOI: 10.1016/j.celrep.2022.110307
Fat3 acts through independent cytoskeletal effectors to coordinate asymmetric cell behaviors during polarized circuit assembly
Abstract
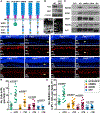
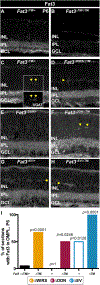
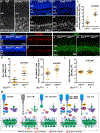
The polarized flow of information through neural circuits depends on the orderly arrangement of neurons, their processes, and their synapses. This polarity emerges sequentially in development, starting with the directed migration of neuronal precursors, which subsequently elaborate neurites that form synapses in specific locations. In other organs, Fat cadherins sense the position and then polarize individual cells by inducing localized changes in the cytoskeleton that are coordinated across the tissue. Here, we show that the Fat-related protein Fat3 plays an analogous role during the assembly of polarized circuits in the murine retina. We find that the Fat3 intracellular domain (ICD) binds to cytoskeletal regulators and synaptic proteins, with discrete motifs required for amacrine cell migration and neurite retraction. Moreover, upon ICD deletion, extra neurites form but do not make ectopic synapses, suggesting that Fat3 independently regulates synapse localization. Thus, Fat3 serves as a molecular node to coordinate asymmetric cell behaviors across development.
Keywords: fat cadherins; neurite retraction; neuronal migration; retinal development; synapse localization.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Avilés EC, Krol A, Henle SJ, Burroughs-Garcia J, Deans MR, and Goodrich LV (2022a). Mass Spectrometry data for: Fat3 acts through independent cytoskeletal effectors to coordinate asymmetric cell behaviors during polarized circuit assembly. Harvard Dataverse, V1. 10.7910/DVN/TXMJYK. - DOI - PMC - PubMed
-
- Badouel C, Zander M, Liscio N, Bagherie-Lachidan M, Sopko R, Coyaud E, Raught B, Miller F, and McNeill H (2015). Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development 142, 2781–2791. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

