LncRNA FIRRE functions as a tumor promoter by interaction with PTBP1 to stabilize BECN1 mRNA and facilitate autophagy
- PMID: 35110535
- PMCID: PMC8811066
- DOI: 10.1038/s41419-022-04509-1
LncRNA FIRRE functions as a tumor promoter by interaction with PTBP1 to stabilize BECN1 mRNA and facilitate autophagy
Abstract
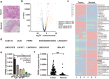
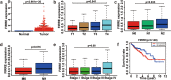
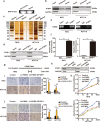
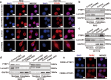
Long non-coding RNAs (lncRNAs) play critical functions in various cancers. Firre intergenic repeating RNA element (FIRRE), a lncRNA located in the nucleus, was overexpressed in colorectal cancer (CRC). However, the detailed mechanism of FIRRE in CRC remains elusive. Results of RNA sequence and qPCR illustrated overexpression of FIRRE in CRC cell lines and tissues. The aberrant expression of FIRRE was correlated with the migration, invasion, and proliferation in cell lines. In accordance, it was also associated with lymphatic metastasis and distant metastasis in patients with CRC. FIRRE was identified to physically interact with Polypyrimidine tract-binding protein (PTBP1) by RNA pull-down and RNA immunoprecipitation (RIP). Overexpression of FIRRE induced the translocation of PTBP1 from nucleus to cytoplasm, which was displayed by immunofluorescence and western blot. In turn, delocalization of FIRRE from nucleus to cytoplasm is observed after the loss of PTBP1. The RNA-protein complex in the cytoplasm directly bound to BECN1 mRNA, and the binding site was at the 3' end of the mRNA. Cells with FIRRE and PTBP1 depletion alone or in combination were treated by Actinomycin D (ACD). Results of qPCR showed FIRRE stabilized BECN1 mRNA in a PTBP1-medieated manner. In addition, FIRRE contributed to autophagy activity. These findings indicate FIRRE acts as an oncogenic factor in CRC, which induces tumor development through stabilizing BECN1 mRNA and facilitating autophagy in a PTBP1-mediated manner.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


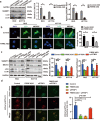

References
-
- Zheng RS, Sun KX, Zhang SW, Zeng HM, Zou XN, Chen R, et al. [Report of cancer epidemiology in China, 2015] Zhonghua Zhong Liu Za Zhi. 2019;41:19–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 15411969800/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
- 15411969800/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
- 15411969800/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
- 15411969800/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
LinkOut - more resources
Full Text Sources
Miscellaneous

