Long non-coding RNA PAARH promotes hepatocellular carcinoma progression and angiogenesis via upregulating HOTTIP and activating HIF-1α/VEGF signaling
- PMID: 35110549
- PMCID: PMC8810756
- DOI: 10.1038/s41419-022-04505-5
Long non-coding RNA PAARH promotes hepatocellular carcinoma progression and angiogenesis via upregulating HOTTIP and activating HIF-1α/VEGF signaling
Abstract
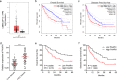
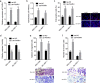
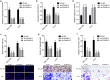
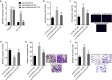
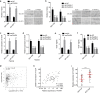
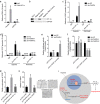
Hepatocellular carcinoma (HCC) is one of the leading lethal malignancies and a hypervascular tumor. Although some long non-coding RNAs (lncRNAs) have been revealed to be involved in HCC. The contributions of lncRNAs to HCC progression and angiogenesis are still largely unknown. In this study, we identified a HCC-related lncRNA, CMB9-22P13.1, which was highly expressed and correlated with advanced stage, vascular invasion, and poor survival in HCC. We named this lncRNA Progression and Angiogenesis Associated RNA in HCC (PAARH). Gain- and loss-of function assays revealed that PAARH facilitated HCC cellular growth, migration, and invasion, repressed HCC cellular apoptosis, and promoted HCC tumor growth and angiogenesis in vivo. PAARH functioned as a competing endogenous RNA to upregulate HOTTIP via sponging miR-6760-5p, miR-6512-3p, miR-1298-5p, miR-6720-5p, miR-4516, and miR-6782-5p. The expression of PAARH was significantly positively associated with HOTTIP in HCC tissues. Functional rescue assays verified that HOTTIP was a critical mediator of the roles of PAARH in modulating HCC cellular growth, apoptosis, migration, and invasion. Furthermore, PAARH was found to physically bind hypoxia inducible factor-1 subunit alpha (HIF-1α), facilitate the recruitment of HIF-1α to VEGF promoter, and activate VEGF expression under hypoxia, which was responsible for the roles of PAARH in promoting angiogenesis. The expression of PAARH was positively associated with VEGF expression and microvessel density in HCC tissues. In conclusion, these findings demonstrated that PAARH promoted HCC progression and angiogenesis via upregulating HOTTIP and activating HIF-1α/VEGF signaling. PAARH represents a potential prognostic biomarker and therapeutic target for HCC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

