Engineered bacterial voltage-gated sodium channel platform for cardiac gene therapy
- PMID: 35110560
- PMCID: PMC8810800
- DOI: 10.1038/s41467-022-28251-6
Engineered bacterial voltage-gated sodium channel platform for cardiac gene therapy
Erratum in
-
Author Correction: Engineered bacterial voltage-gated sodium channel platform for cardiac gene therapy.Nat Commun. 2023 Nov 16;14(1):7411. doi: 10.1038/s41467-023-43260-9. Nat Commun. 2023. PMID: 37973962 Free PMC article. No abstract available.
Abstract
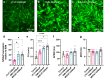
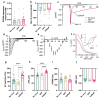
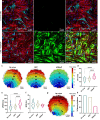
Therapies for cardiac arrhythmias could greatly benefit from approaches to enhance electrical excitability and action potential conduction in the heart by stably overexpressing mammalian voltage-gated sodium channels. However, the large size of these channels precludes their incorporation into therapeutic viral vectors. Here, we report a platform utilizing small-size, codon-optimized engineered prokaryotic sodium channels (BacNav) driven by muscle-specific promoters that significantly enhance excitability and conduction in rat and human cardiomyocytes in vitro and adult cardiac tissues from multiple species in silico. We also show that the expression of BacNav significantly reduces occurrence of conduction block and reentrant arrhythmias in fibrotic cardiac cultures. Moreover, functional BacNav channels are stably expressed in healthy mouse hearts six weeks following intravenous injection of self-complementary adeno-associated virus (scAAV) without causing any adverse effects on cardiac electrophysiology. The large diversity of prokaryotic sodium channels and experimental-computational platform reported in this study should facilitate the development and evaluation of BacNav-based gene therapies for cardiac conduction disorders.
© 2022. The Author(s).
Conflict of interest statement
H.X.N., T.W., and N.B. are inventors on a pending patent concerning application of prokaryotic sodium channels (PCT:WO2021076600A1). The remaining authors declare no competing interests.
Figures





Similar articles
-
Prokaryotic voltage-gated sodium channels are more effective than endogenous Nav1.5 channels in rescuing cardiac action potential conduction: an in silico study.Am J Physiol Heart Circ Physiol. 2023 Nov 1;325(5):H1178-H1192. doi: 10.1152/ajpheart.00287.2023. Epub 2023 Sep 22. Am J Physiol Heart Circ Physiol. 2023. PMID: 37737736 Free PMC article.
-
Engineering prokaryotic channels for control of mammalian tissue excitability.Nat Commun. 2016 Oct 18;7:13132. doi: 10.1038/ncomms13132. Nat Commun. 2016. PMID: 27752065 Free PMC article.
-
Cardiac Kir2.1 and NaV1.5 Channels Traffic Together to the Sarcolemma to Control Excitability.Circ Res. 2018 May 25;122(11):1501-1516. doi: 10.1161/CIRCRESAHA.117.311872. Epub 2018 Mar 7. Circ Res. 2018. PMID: 29514831 Free PMC article.
-
Sodium channel (dys)function and cardiac arrhythmias.Cardiovasc Ther. 2010 Oct;28(5):287-94. doi: 10.1111/j.1755-5922.2010.00210.x. Epub 2010 Jul 14. Cardiovasc Ther. 2010. PMID: 20645984 Review.
-
Heritable arrhythmia syndromes associated with abnormal cardiac sodium channel function: ionic and non-ionic mechanisms.Cardiovasc Res. 2020 Jul 15;116(9):1557-1570. doi: 10.1093/cvr/cvaa082. Cardiovasc Res. 2020. PMID: 32251506 Free PMC article. Review.
Cited by
-
Microbial membrane transport proteins and their biotechnological applications.World J Microbiol Biotechnol. 2024 Jan 16;40(2):71. doi: 10.1007/s11274-024-03891-6. World J Microbiol Biotechnol. 2024. PMID: 38225445 Free PMC article. Review.
-
The Genetic Basis of Sudden Cardiac Death: From Diagnosis to Emerging Genetic Therapies.Annu Rev Med. 2025 Jan;76(1):283-299. doi: 10.1146/annurev-med-042423-042903. Epub 2025 Jan 16. Annu Rev Med. 2025. PMID: 39499917 Review.
-
Insights into the prospects of nanobiomaterials in the treatment of cardiac arrhythmia.J Nanobiotechnology. 2024 Aug 30;22(1):523. doi: 10.1186/s12951-024-02805-w. J Nanobiotechnology. 2024. PMID: 39215361 Free PMC article. Review.
-
Inhalable CAR-T cell-derived exosomes as paclitaxel carriers for treating lung cancer.J Transl Med. 2023 Jun 12;21(1):383. doi: 10.1186/s12967-023-04206-3. J Transl Med. 2023. PMID: 37308954 Free PMC article.
-
BacNav Gene Therapy Improves Function of Infarcted Engineered Human Myocardium and NHPs.Circ Res. 2025 Aug 29;137(6):899-901. doi: 10.1161/CIRCRESAHA.125.326570. Epub 2025 Aug 1. Circ Res. 2025. PMID: 40747546 Free PMC article. No abstract available.
References
-
- Janse MJ, Wit AL. Electrophysiological mechanisms of ventricular arrhythmias resulting from myocardial ischemia and infarction. Physiol. Rev. 1989;69:1049–1169. - PubMed
-
- Carmeliet E. Cardiac ionic currents and acute ischemia: from channels to arrhythmias. Physiol. Rev. 1999;79:917–1017. - PubMed
-
- Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

