Preharvest UV-C Hormesis Induces Key Genes Associated With Homeostasis, Growth and Defense in Lettuce Inoculated With Xanthomonas campestris pv. vitians
- PMID: 35111177
- PMCID: PMC8801786
- DOI: 10.3389/fpls.2021.793989
Preharvest UV-C Hormesis Induces Key Genes Associated With Homeostasis, Growth and Defense in Lettuce Inoculated With Xanthomonas campestris pv. vitians
Abstract
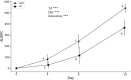
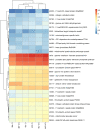
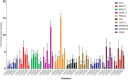
Preharvest application of hormetic doses of ultraviolet-C (UV-C) generates beneficial effects in plants. In this study, within 1 week, four UV-C treatments of 0.4 kJ/m2 were applied to 3-week-old lettuce seedlings. The leaves were inoculated with a virulent strain of Xanthomonas campestris pv. vitians (Xcv) 48 h after the last UV-C application. The extent of the disease was tracked over time and a transcriptomic analysis was performed on lettuce leaf samples. Samples of lettuce leaves, from both control and treated groups, were taken at two different times corresponding to T2, 48 h after the last UV-C treatment and T3, 24 h after inoculation (i.e., 72 h after the last UV-C treatment). A significant decrease in disease severity between the UV-C treated lettuce and the control was observed on days 4, 8, and 14 after pathogen inoculation. Data from the transcriptomic study revealed, that in response to the effect of UV-C alone and/or UV-C + Xcv, a total of 3828 genes were differentially regulated with fold change (|log2-FC|) > 1.5 and false discovery rate (FDR) < 0.05. Among these, of the 2270 genes of known function 1556 were upregulated and 714 were downregulated. A total of 10 candidate genes were verified by qPCR and were generally consistent with the transcriptomic results. The differentially expressed genes observed in lettuce under the conditions of the present study were associated with 14 different biological processes in the plant. These genes are involved in a series of metabolic pathways associated with the ability of lettuce treated with hormetic doses of UV-C to resume normal growth and to defend themselves against potential stressors. The results indicate that the hormetic dose of UV-C applied preharvest on lettuce in this study, can be considered as an eustress that does not interfere with the ability of the treated plants to carry on a set of key physiological processes namely: homeostasis, growth and defense.
Keywords: bacterial disease; cell homeostasis; defense mechanisms; eustress; leaf spot disease; lettuce; physiological processes; plant growth.
Copyright © 2022 Sidibé, Charles, Lucier, Xu and Beaulieu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Bacterial Leaf Spot of Iceberg Lettuce Caused by Xanthomonas campestris pv. vitians Type B, a New Disease in South Korea.Plant Dis. 2010 Jun;94(6):790. doi: 10.1094/PDIS-94-6-0790B. Plant Dis. 2010. PMID: 30754343
-
First Report of Bacterial Leaf Spot of Lettuce (Lactuca sativa) Caused by Xanthomonas campestris pv. vitians in Saudi Arabia.Plant Dis. 2009 Jan;93(1):107. doi: 10.1094/PDIS-93-1-0107B. Plant Dis. 2009. PMID: 30764273
-
Bacterial Leaf Spot of Lettuce: Relationship of Temperature to Infection and Potential Host Range of Xanthomonas campestris pv. vitians.Plant Dis. 2006 Apr;90(4):465-470. doi: 10.1094/PD-90-0465. Plant Dis. 2006. PMID: 30786595
-
Role of Crop Debris and Weeds in the Epidemiology of Bacterial Leaf Spot of Lettuce in California.Plant Dis. 2001 Feb;85(2):169-178. doi: 10.1094/PDIS.2001.85.2.169. Plant Dis. 2001. PMID: 30831938
-
A Comprehensive Overview of the Genes and Functions Required for Lettuce Infection by the Hemibiotrophic Phytopathogen Xanthomonas hortorum pv. vitians.mSystems. 2022 Apr 26;7(2):e0129021. doi: 10.1128/msystems.01290-21. Epub 2022 Mar 21. mSystems. 2022. PMID: 35311560 Free PMC article. Review.
Cited by
-
Prominent Anti-UVC Activity of Lignin Degradation Products.In Vivo. 2022 Nov-Dec;36(6):2689-2699. doi: 10.21873/invivo.13004. In Vivo. 2022. PMID: 36309360 Free PMC article.
-
Comparison of UVC Sensitivity and Dectin-2 Expression Between Malignant and Non-malignant Cells.In Vivo. 2022 Sep-Oct;36(5):2116-2125. doi: 10.21873/invivo.12937. In Vivo. 2022. PMID: 36099100 Free PMC article.
References
-
- Achuo E. A., Prinsen E., Höfte M. (2006). Influence of drought, salt stress and abscisic acid on the resistance of tomato to Botrytis cinerea and Oidium neolycopersici. Plant Pathol. 55 178–186. 10.1111/j.1365-3059.2006.01340.x - DOI
-
- Bagati S., Mahajan R., Nazir M., Dar A. A., Zargar S. M. (2018). “‘Omics’: a gateway towards abiotic stress tolerance,” in Abiotic Stress-Mediated Sensing and Signaling in Plants: An Omics Perspective, eds Zargar S., Zargar M. (Singapore: Springer; ), 1–45. 10.1007/978-981-10-7479-0_1 - DOI
-
- Balagué C., Gouget A., Bouchez O., Souriac C., Haget N., Boutet-Mercey S., et al. (2017). The Arabidopsis thaliana lectin receptor kinase LecRK-I. 9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 18 937–948. 10.1111/mpp.12457 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

