DnoisE: distance denoising by entropy. An open-source parallelizable alternative for denoising sequence datasets
- PMID: 35111399
- PMCID: PMC8783565
- DOI: 10.7717/peerj.12758
DnoisE: distance denoising by entropy. An open-source parallelizable alternative for denoising sequence datasets
Abstract
DNA metabarcoding is broadly used in biodiversity studies encompassing a wide range of organisms. Erroneous amplicons, generated during amplification and sequencing procedures, constitute one of the major sources of concern for the interpretation of metabarcoding results. Several denoising programs have been implemented to detect and eliminate these errors. However, almost all denoising software currently available has been designed to process non-coding ribosomal sequences, most notably prokaryotic 16S rDNA. The growing number of metabarcoding studies using coding markers such as COI or RuBisCO demands a re-assessment and calibration of denoising algorithms. Here we present DnoisE, the first denoising program designed to detect erroneous reads and merge them with the correct ones using information from the natural variability (entropy) associated to each codon position in coding barcodes. We have developed an open-source software using a modified version of the UNOISE algorithm. DnoisE implements different merging procedures as options, and can incorporate codon entropy information either retrieved from the data or supplied by the user. In addition, the algorithm of DnoisE is parallelizable, greatly reducing runtimes on computer clusters. Our program also allows different input file formats, so it can be readily incorporated into existing metabarcoding pipelines.
Keywords: Bioinformatic pipelines; Coding markers; Denoising algorithms; Entropy correction; Metabarcoding; Metaphylogeography.
©2022 Antich et al.
Conflict of interest statement
Owen S. Wangensteen is an Academic Editor for PeerJ.
Figures


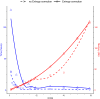
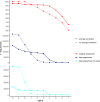
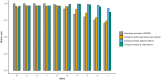
References
-
- Brandt MI, Trouche B, Quintric L, Günther B, Wincker P, Poulain J, Arnaud-Haond S. Bioinformatic pipelines combining denoising and clustering tools allow for more comprehensive prokaryotic and eukaryotic metabarcoding. Molecular Ecology Resources. 2021;21(6):1904–1921. doi: 10.1111/1755-099813398. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

