Transcriptomic and Metabolomic Analyses Reveal Inhibition of Hepatic Adipogenesis and Fat Catabolism in Yak for Adaptation to Forage Shortage During Cold Season
- PMID: 35111749
- PMCID: PMC8802892
- DOI: 10.3389/fcell.2021.759521
Transcriptomic and Metabolomic Analyses Reveal Inhibition of Hepatic Adipogenesis and Fat Catabolism in Yak for Adaptation to Forage Shortage During Cold Season
Abstract
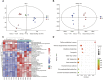
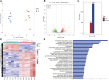
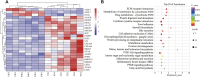
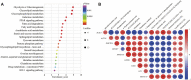
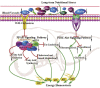
Animals have adapted behavioral and physiological strategies to conserve energy during periods of adverse conditions. Hepatic glucose is one such adaptation used by grazing animals. While large vertebrates have been shown to have feed utilization and deposition of nutrients-fluctuations in metabolic rate-little is known about the regulating mechanism that controls hepatic metabolism in yaks under grazing conditions in the cold season. Hence, the objective of this research was to integrate transcriptomic and metabolomic data to better understand how the hepatic responds to chronic nutrient stress. Our analyses indicated that the blood parameters related to energy metabolism (glucose, total cholesterol, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, lipoprotein lipase, insulin, and insulin-like growth factor 1) were significantly (p < 0.05) lower in the cold season. The RNA-Seq results showed that malnutrition inhibited lipid synthesis (particularly fatty acid, cholesterol, and steroid synthesis), fatty acid oxidation, and lipid catabolism and promoted gluconeogenesis by inhibiting the peroxisome proliferator-activated receptor (PPAR) and PI3K-Akt signaling pathways. For metabolite profiles, 359 metabolites were significantly altered in two groups. Interestingly, the cold season group remarkably decreased glutathione and phosphatidylcholine (18:2 (2E, 4E)/0:0). Moreover, integrative analysis of the transcriptome and metabolome demonstrated that glycolysis or gluconeogenesis, PPAR signaling pathway, fatty acid biosynthesis, steroid biosynthesis, and glutathione metabolism play an important role in the potential relationship between differential expression genes and metabolites. The reduced lipid synthesis, fatty acid oxidation, and fat catabolism facilitated gluconeogenesis by inhibiting the PPAR and PI3K-Akt signaling pathways to maintain the energy homeostasis of the whole body in the yak, thereby coping with the shortage of forages and adapting to the extreme environment of the Qinghai-Tibetan Plateau (QTP).
Keywords: energy metabolism; forage shortage; liver; metabolomics; transcriptome; yak.
Copyright © 2022 Zheng, Du, Zhang, Liang, Ahmad, Shen, Salekdeh and Ding.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Anders S., Huber W. (2012). Differential Expression of RNA-Seq Data at the Gene Level–The DESeq Package, 10. Heidelberg, Germany: European Molecular Biology Laboratory EMBL, f1000.
LinkOut - more resources
Full Text Sources

