Pediatric sepsis phenotypes for enhanced therapeutics: An application of clustering to electronic health records
- PMID: 35112102
- PMCID: PMC8790108
- DOI: 10.1002/emp2.12660
Pediatric sepsis phenotypes for enhanced therapeutics: An application of clustering to electronic health records
Abstract
Objective: The heterogeneity of pediatric sepsis patients suggests the potential benefits of clustering analytics to derive phenotypes with distinct host response patterns that may help guide personalized therapeutics. We evaluate the relative performance of latent class analysis (LCA) and K-means, 2 commonly used clustering methods toward the derivation of clinically useful pediatric sepsis phenotypes.
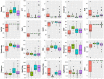
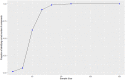
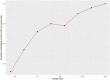
Methods: Data were extracted from anonymized medical records of 6446 pediatric patients that presented to 1 of 6 emergency departments (EDs) between 2013 and 2018 and were thereafter admitted. Using International Classification of Diseases (ICD)-9 and ICD-10 discharge codes, 151 patients were identified with a sepsis continuum diagnosis that included septicemia, sepsis, severe sepsis, and septic shock. Using feature sets used in related clustering studies, LCA and K-means algorithms were used to derive 4 distinct phenotypic pediatric sepsis segmentations. Each segmentation was evaluated for phenotypic homogeneity, separation, and clinical use.
Results: Using the 2 feature sets, LCA clustering resulted in 2 similar segmentations of 4 clinically distinct phenotypes, while K-means clustering resulted in segmentations of 3 and 4 phenotypes. All 4 segmentations identified at least 1 high severity phenotype, but LCA-identified phenotypes reflected superior stratification, high entropy approaching 1 (eg, 0.994) indicating excellent separation between estimated phenotypes, and differential treatment/treatment response, and outcomes that were non-randomly distributed across phenotypes (P < 0.001).
Conclusion: Compared to K-means, which is commonly used in clustering studies, LCA appears to be a more robust, clinically useful statistical tool in analyzing a heterogeneous pediatric sepsis cohort toward informing targeted therapies. Additional prospective studies are needed to validate clinical utility of predictive models that target derived pediatric sepsis phenotypes in emergency department settings.
Keywords: K‐means; LCA; phenotypes; sepsis.
© 2022 The Authors. JACEP Open published by Wiley Periodicals LLC on behalf of American College of Emergency Physicians.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis.JAMA. 2019 May 28;321(20):2003-2017. doi: 10.1001/jama.2019.5791. JAMA. 2019. PMID: 31104070 Free PMC article.
-
Derivation, validation, and transcriptomic assessment of pediatric septic shock phenotypes identified through latent profile analyses: Results from a prospective multi-center observational cohort.Res Sq [Preprint]. 2023 Dec 6:rs.3.rs-3692289. doi: 10.21203/rs.3.rs-3692289/v1. Res Sq. 2023. PMID: 38105983 Free PMC article. Preprint.
-
Derivation and Initial Validation of Clinical Phenotypes of Children Presenting with Concussion Acutely in the Emergency Department: Latent Class Analysis of a Multi-Center, Prospective Cohort, Observational Study.J Neurotrauma. 2019 Jun;36(11):1758-1767. doi: 10.1089/neu.2018.6009. Epub 2019 Mar 6. J Neurotrauma. 2019. PMID: 30618356
-
Clinical Decision-Support Systems for Detection of Systemic Inflammatory Response Syndrome, Sepsis, and Septic Shock in Critically Ill Patients: A Systematic Review.Methods Inf Med. 2019 Dec;58(S 02):e43-e57. doi: 10.1055/s-0039-1695717. Epub 2019 Sep 9. Methods Inf Med. 2019. PMID: 31499571
-
Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012.Crit Care Med. 2013 Feb;41(2):580-637. doi: 10.1097/CCM.0b013e31827e83af. Crit Care Med. 2013. PMID: 23353941
Cited by
-
Balanced crystalloid versus saline for resuscitation in pediatric septic shock: a systematic review and meta-analysis.BMC Pediatr. 2025 Jan 31;25(1):81. doi: 10.1186/s12887-025-05442-w. BMC Pediatr. 2025. PMID: 39891147 Free PMC article.
-
Subgroup identification-based model selection to improve the predictive performance of individualized dosing.J Pharmacokinet Pharmacodyn. 2024 Jun;51(3):253-263. doi: 10.1007/s10928-024-09909-8. Epub 2024 Feb 24. J Pharmacokinet Pharmacodyn. 2024. PMID: 38400995
-
Identification and Prediction of Clinical Phenotypes in Hospitalized Patients With COVID-19: Machine Learning From Medical Records.JMIR Form Res. 2023 Oct 6;7:e46807. doi: 10.2196/46807. JMIR Form Res. 2023. PMID: 37642512 Free PMC article.
-
Clinical Sepsis Phenotypes in Critically Ill Patients.Microorganisms. 2023 Aug 27;11(9):2165. doi: 10.3390/microorganisms11092165. Microorganisms. 2023. PMID: 37764009 Free PMC article. Review.
-
Patient feasibility as a novel approach for integrating IRT and LCA statistical models into patient-centric qualitative data-a pilot study.Front Digit Health. 2024 Oct 2;6:1378497. doi: 10.3389/fdgth.2024.1378497. eCollection 2024. Front Digit Health. 2024. PMID: 39416519 Free PMC article.
References
-
- Oberski D. Mixture Models: Latent Profile and Latent Class Analysis. In: Robertson J., Kaptein M. (eds) Modern Statistical Methods for HCI. Human Computer Interaction Series. Springer, Cham. 2016. 10.1007/978-3-319-26633-6_12 - DOI
-
- Boehmke B, Greenwell B. Hands‐On Machine Learning with R (1st ed.). Chapman and Hall/CRC. (2019). 10.1201/9780367816377 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
