High-Performance Deep Learning Toolbox for Genome-Scale Prediction of Protein Structure and Function
- PMID: 35112110
- PMCID: PMC8802329
- DOI: 10.1109/mlhpc54614.2021.00010
High-Performance Deep Learning Toolbox for Genome-Scale Prediction of Protein Structure and Function
Abstract
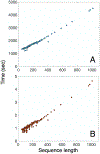
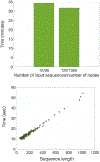
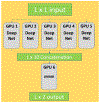
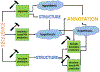
Computational biology is one of many scientific disciplines ripe for innovation and acceleration with the advent of high-performance computing (HPC). In recent years, the field of machine learning has also seen significant benefits from adopting HPC practices. In this work, we present a novel HPC pipeline that incorporates various machine-learning approaches for structure-based functional annotation of proteins on the scale of whole genomes. Our pipeline makes extensive use of deep learning and provides computational insights into best practices for training advanced deep-learning models for high-throughput data such as proteomics data. We showcase methodologies our pipeline currently supports and detail future tasks for our pipeline to envelop, including large-scale sequence comparison using SAdLSA and prediction of protein tertiary structures using AlphaFold2.
Keywords: computational biology; deep learning; high-performance computing; machine learning; protein sequence alignment; protein structure prediction.
Figures
Similar articles
-
A General Framework to Learn Tertiary Structure for Protein Sequence Characterization.Front Bioinform. 2021 May;1:689960. doi: 10.3389/fbinf.2021.689960. Epub 2021 May 21. Front Bioinform. 2021. PMID: 34308415 Free PMC article.
-
A novel sequence alignment algorithm based on deep learning of the protein folding code.Bioinformatics. 2021 May 1;37(4):490-496. doi: 10.1093/bioinformatics/btaa810. Bioinformatics. 2021. PMID: 32960943 Free PMC article.
-
DeepBIO: an automated and interpretable deep-learning platform for high-throughput biological sequence prediction, functional annotation and visualization analysis.Nucleic Acids Res. 2023 Apr 24;51(7):3017-3029. doi: 10.1093/nar/gkad055. Nucleic Acids Res. 2023. PMID: 36796796 Free PMC article.
-
Protein Structure Prediction: Conventional and Deep Learning Perspectives.Protein J. 2021 Aug;40(4):522-544. doi: 10.1007/s10930-021-10003-y. Epub 2021 May 28. Protein J. 2021. PMID: 34050498 Review.
-
Data Integration Using Advances in Machine Learning in Drug Discovery and Molecular Biology.Methods Mol Biol. 2021;2190:167-184. doi: 10.1007/978-1-0716-0826-5_7. Methods Mol Biol. 2021. PMID: 32804365 Review.
Cited by
-
De Novo Atomic Protein Structure Modeling for Cryo-EM Density Maps Using 3D Transformer and Hidden Markov Model.bioRxiv [Preprint]. 2024 Jan 2:2024.01.02.573943. doi: 10.1101/2024.01.02.573943. bioRxiv. 2024. Update in: Nat Commun. 2024 Jun 29;15(1):5511. doi: 10.1038/s41467-024-49647-6. PMID: 38260535 Free PMC article. Updated. Preprint.
-
De novo atomic protein structure modeling for cryoEM density maps using 3D transformer and HMM.Nat Commun. 2024 Jun 29;15(1):5511. doi: 10.1038/s41467-024-49647-6. Nat Commun. 2024. PMID: 38951555 Free PMC article.
-
AF2Complex predicts direct physical interactions in multimeric proteins with deep learning.Nat Commun. 2022 Apr 1;13(1):1744. doi: 10.1038/s41467-022-29394-2. Nat Commun. 2022. PMID: 35365655 Free PMC article.
-
Multi-head attention-based U-Nets for predicting protein domain boundaries using 1D sequence features and 2D distance maps.BMC Bioinformatics. 2022 Jul 19;23(1):283. doi: 10.1186/s12859-022-04829-1. BMC Bioinformatics. 2022. PMID: 35854211 Free PMC article.
-
Cryo2StructData: A Large Labeled Cryo-EM Density Map Dataset for AI-based Modeling of Protein Structures.bioRxiv [Preprint]. 2024 Jan 2:2023.06.14.545024. doi: 10.1101/2023.06.14.545024. bioRxiv. 2024. Update in: Sci Data. 2024 May 6;11(1):458. doi: 10.1038/s41597-024-03299-9. PMID: 37398020 Free PMC article. Updated. Preprint.
References
-
- Baines Mandeep, Bhosale Shruti, Caggiano Vittorio, Goyal Naman, Goyal Siddharth, Ott Myle, Lefaudeux Benjamin, Liptchinsky Vitaliy, Rabbat Mike, Sheiffer Sam, Sridhar Anjali, and Xu Min. Fairscale: A general purpose modular pytorch library for high performance and large scale training.
-
- Biewald Lukas. Experiment tracking with weights and biases, 2020. Software available from wandb.com.
-
- Brown Tom B, Mann Benjamin, Ryder Nick, Subbiah Melanie, Kaplan Jared, Dhariwal Prafulla, Neelakantan Arvind, Shyam Pranav, Sastry Girish, Askell Amanda, et al. Language models are few-shot learners. arXiv preprint arXiv:2005.14165, 2020.
-
- Chen Chen, Chen Xiao, Wu Tianqi, Alex Morehead, and Cheng Jianlin. Improved protein structure accuracy estimation with graph-based equivariant networks. In preparation, 2021.
Grants and funding
LinkOut - more resources
Full Text Sources
