High-Performance Deep Learning Toolbox for Genome-Scale Prediction of Protein Structure and Function
- PMID: 35112110
- PMCID: PMC8802329
- DOI: 10.1109/mlhpc54614.2021.00010
High-Performance Deep Learning Toolbox for Genome-Scale Prediction of Protein Structure and Function
Abstract
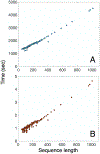
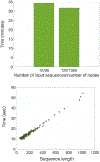
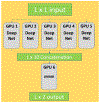
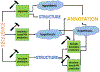
Computational biology is one of many scientific disciplines ripe for innovation and acceleration with the advent of high-performance computing (HPC). In recent years, the field of machine learning has also seen significant benefits from adopting HPC practices. In this work, we present a novel HPC pipeline that incorporates various machine-learning approaches for structure-based functional annotation of proteins on the scale of whole genomes. Our pipeline makes extensive use of deep learning and provides computational insights into best practices for training advanced deep-learning models for high-throughput data such as proteomics data. We showcase methodologies our pipeline currently supports and detail future tasks for our pipeline to envelop, including large-scale sequence comparison using SAdLSA and prediction of protein tertiary structures using AlphaFold2.
Keywords: computational biology; deep learning; high-performance computing; machine learning; protein sequence alignment; protein structure prediction.
Figures
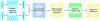
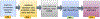
References
-
- Baines Mandeep, Bhosale Shruti, Caggiano Vittorio, Goyal Naman, Goyal Siddharth, Ott Myle, Lefaudeux Benjamin, Liptchinsky Vitaliy, Rabbat Mike, Sheiffer Sam, Sridhar Anjali, and Xu Min. Fairscale: A general purpose modular pytorch library for high performance and large scale training.
-
- Biewald Lukas. Experiment tracking with weights and biases, 2020. Software available from wandb.com.
-
- Brown Tom B, Mann Benjamin, Ryder Nick, Subbiah Melanie, Kaplan Jared, Dhariwal Prafulla, Neelakantan Arvind, Shyam Pranav, Sastry Girish, Askell Amanda, et al. Language models are few-shot learners. arXiv preprint arXiv:2005.14165, 2020.
-
- Chen Chen, Chen Xiao, Wu Tianqi, Alex Morehead, and Cheng Jianlin. Improved protein structure accuracy estimation with graph-based equivariant networks. In preparation, 2021.
Grants and funding
LinkOut - more resources
Full Text Sources
