Systems pharmacology-based drug discovery and active mechanism of natural products for coronavirus pneumonia (COVID-19): An example using flavonoids
- PMID: 35114443
- PMCID: PMC8789666
- DOI: 10.1016/j.compbiomed.2022.105241
Systems pharmacology-based drug discovery and active mechanism of natural products for coronavirus pneumonia (COVID-19): An example using flavonoids
Abstract
Background: Recently, the value of natural products has been extensively considered because these resources can potentially be applied to prevent and treat coronavirus pneumonia 2019 (COVID-19). However, the discovery of nature drugs is problematic because of their complex composition and active mechanisms.
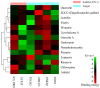
Methods: This comprehensive study was performed on flavonoids, which are compounds with anti-inflammatory and antiviral effects, to show drug discovery and active mechanism from natural products in the treatment of COVID-19 via a systems pharmacological model. First, a chemical library of 255 potential flavonoids was constructed. Second, the pharmacodynamic basis and mechanism of action between flavonoids and COVID-19 were explored by constructing a compound-target and target-disease network, targets protein-protein interaction (PPI), MCODE analysis, gene ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment.
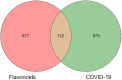
Results: In total, 105 active flavonoid components were identified, of which 6 were major candidate compounds (quercetin, epigallocatechin-3-gallate (EGCG), luteolin, fisetin, wogonin, and licochalcone A). 152 associated targets were yielded based on network construction, and 7 family proteins (PTGS, GSK3β, ABC, NOS, EGFR, and IL) were included as central hub targets. Moreover, 528 GO items and 178 KEGG pathways were selected through enrichment of target functions. Lastly, molecular docking demonstrated good stability of the combination of selected flavonoids with 3CL Pro and ACEⅡ.
Conclusion: Natural flavonoids could enable resistance against COVID-19 by regulating inflammatory, antiviral, and immune responses, and repairing tissue injury. This study has scientific significance for the selective utilization of natural products, medicinal value enhancement of flavonoids, and drug screening for the treatment of COVID-19 induced by SARS-COV-2.
Keywords: COVID-19; Flavonoids; Molecular docking; Natural products; Systems pharmacology.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures








Similar articles
-
Identification of phytochemical compounds of Fagopyrum dibotrys and their targets by metabolomics, network pharmacology and molecular docking studies.Heliyon. 2023 Mar;9(3):e14029. doi: 10.1016/j.heliyon.2023.e14029. Epub 2023 Mar 1. Heliyon. 2023. PMID: 36911881 Free PMC article.
-
Network pharmacology, molecular docking integrated surface plasmon resonance technology reveals the mechanism of Toujie Quwen Granules against coronavirus disease 2019 pneumonia.Phytomedicine. 2021 May;85:153401. doi: 10.1016/j.phymed.2020.153401. Epub 2020 Oct 28. Phytomedicine. 2021. PMID: 33191068 Free PMC article.
-
Potential mechanism prediction of Cold-Damp Plague Formula against COVID-19 via network pharmacology analysis and molecular docking.Chin Med. 2020 Jul 30;15:78. doi: 10.1186/s13020-020-00360-8. eCollection 2020. Chin Med. 2020. PMID: 32754224 Free PMC article.
-
Network pharmacology and molecular docking analyses on Lianhua Qingwen capsule indicate Akt1 is a potential target to treat and prevent COVID-19.Cell Prolif. 2020 Dec;53(12):e12949. doi: 10.1111/cpr.12949. Epub 2020 Nov 3. Cell Prolif. 2020. PMID: 33140889 Free PMC article.
-
The mechanism and active compounds of semen armeniacae amarum treating coronavirus disease 2019 based on network pharmacology and molecular docking.Food Nutr Res. 2021 Feb 4;65. doi: 10.29219/fnr.v65.5623. eCollection 2021. Food Nutr Res. 2021. PMID: 34908920 Free PMC article.
Cited by
-
Commercially Available Flavonols Are Better SARS-CoV-2 Inhibitors than Isoflavone and Flavones.Viruses. 2022 Jun 30;14(7):1458. doi: 10.3390/v14071458. Viruses. 2022. PMID: 35891437 Free PMC article.
-
Phase I/II clinical trial of efficacy and safety of EGCG oxygen nebulization inhalation in the treatment of COVID-19 pneumonia patients with cancer.BMC Cancer. 2024 Apr 17;24(1):486. doi: 10.1186/s12885-024-12228-3. BMC Cancer. 2024. PMID: 38632501 Free PMC article. Clinical Trial.
-
Phenolic compounds versus SARS-CoV-2: An update on the main findings against COVID-19.Heliyon. 2022 Sep 20;8(9):e10702. doi: 10.1016/j.heliyon.2022.e10702. eCollection 2022 Sep. Heliyon. 2022. PMID: 36157310 Free PMC article. Review.
-
Zirconium-Based Metal-Organic Frameworks as Acriflavine Cargos in the Battle against Coronaviruses─A Theoretical and Experimental Approach.ACS Appl Mater Interfaces. 2022 Jun 29;14(25):28615-28627. doi: 10.1021/acsami.2c06420. Epub 2022 Jun 14. ACS Appl Mater Interfaces. 2022. PMID: 35700479 Free PMC article.
-
Endophytic Fungi Isolated from Ageratina adenophora Exhibits Potential Antimicrobial Activity against Multidrug-Resistant Staphylococcus aureus.Plants (Basel). 2023 Feb 1;12(3):650. doi: 10.3390/plants12030650. Plants (Basel). 2023. PMID: 36771733 Free PMC article.
References
-
- World Health Organization, WHO Coronavirus (COVID-19) Dashboard, https://covid19.who.int/, (accessed January 2022).
-
- National Health Commission of the People’s Republic of China, Notice On Issuing The Diagnosis And Treatment Protocol For Novel Coronavirus Pneumonia, http://www. nhc.gov.cn/xcs/xxgzbd/gzbd_index.shtml, (accessed January 2022).
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

