Mitochondrial protein import determines lifespan through metabolic reprogramming and de novo serine biosynthesis
- PMID: 35115503
- PMCID: PMC8814026
- DOI: 10.1038/s41467-022-28272-1
Mitochondrial protein import determines lifespan through metabolic reprogramming and de novo serine biosynthesis
Abstract
Sustained mitochondrial fitness relies on coordinated biogenesis and clearance. Both processes are regulated by constant targeting of proteins into the organelle. Thus, mitochondrial protein import sets the pace for mitochondrial abundance and function. However, our understanding of mitochondrial protein translocation as a regulator of longevity remains enigmatic. Here, we targeted the main protein import translocases and assessed their contribution to mitochondrial abundance and organismal physiology. We find that reduction in cellular mitochondrial load through mitochondrial protein import system suppression, referred to as MitoMISS, elicits a distinct longevity paradigm. We show that MitoMISS triggers the mitochondrial unfolded protein response, orchestrating an adaptive reprogramming of metabolism. Glycolysis and de novo serine biosynthesis are causatively linked to longevity, whilst mitochondrial chaperone induction is dispensable for lifespan extension. Our findings extent the pro-longevity role of UPRmt and provide insight, relevant to the metabolic alterations that promote or undermine survival and longevity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



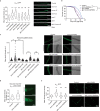


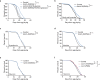
Comment in
-
Shifting metabolism to increase lifespan.Trends Endocrinol Metab. 2022 Aug;33(8):533-535. doi: 10.1016/j.tem.2022.05.001. Epub 2022 May 19. Trends Endocrinol Metab. 2022. PMID: 35597712
Similar articles
-
Compromised Mitochondrial Protein Import Acts as a Signal for UPRmt.Cell Rep. 2019 Aug 13;28(7):1659-1669.e5. doi: 10.1016/j.celrep.2019.07.049. Cell Rep. 2019. PMID: 31412237
-
The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation?Exp Gerontol. 2014 Aug;56:142-6. doi: 10.1016/j.exger.2014.02.002. Epub 2014 Feb 8. Exp Gerontol. 2014. PMID: 24518875 Free PMC article. Review.
-
Effects of the mitochondrial respiratory chain on longevity in C. elegans.Exp Gerontol. 2014 Aug;56:245-55. doi: 10.1016/j.exger.2014.03.028. Epub 2014 Apr 5. Exp Gerontol. 2014. PMID: 24709342 Review.
-
The UPRmt preserves mitochondrial import to extend lifespan.J Cell Biol. 2022 Jul 4;221(7):e202201071. doi: 10.1083/jcb.202201071. Epub 2022 May 24. J Cell Biol. 2022. PMID: 35608535 Free PMC article.
-
Signaling the mitochondrial unfolded protein response.Biochim Biophys Acta. 2013 Feb;1833(2):410-6. doi: 10.1016/j.bbamcr.2012.02.019. Epub 2012 Mar 14. Biochim Biophys Acta. 2013. PMID: 22445420 Free PMC article. Review.
Cited by
-
Mitochondrial protein import stress.Nat Cell Biol. 2025 Feb;27(2):188-201. doi: 10.1038/s41556-024-01590-w. Epub 2025 Jan 22. Nat Cell Biol. 2025. PMID: 39843636 Review.
-
Serine metabolism in macrophage polarization.Inflamm Res. 2024 Jan;73(1):83-98. doi: 10.1007/s00011-023-01815-y. Epub 2023 Dec 9. Inflamm Res. 2024. PMID: 38070057 Review.
-
C. elegans as an Animal Model to Study the Intersection of DNA Repair, Aging and Neurodegeneration.Front Aging. 2022 Jun 22;3:916118. doi: 10.3389/fragi.2022.916118. eCollection 2022. Front Aging. 2022. PMID: 35821838 Free PMC article. Review.
-
Metformin modulates the unfolded protein responses, altering lifespan and health-promoting effects in UPR-activated worms.PLoS One. 2025 Jun 16;20(6):e0326100. doi: 10.1371/journal.pone.0326100. eCollection 2025. PLoS One. 2025. PMID: 40522955 Free PMC article.
-
Mitochondrial Stress and Mitokines: Therapeutic Perspectives for the Treatment of Metabolic Diseases.Diabetes Metab J. 2024 Jan;48(1):1-18. doi: 10.4093/dmj.2023.0115. Epub 2024 Jan 3. Diabetes Metab J. 2024. PMID: 38173375 Free PMC article. Review.
References
-
- Luo C, et al. Mitochondrial accumulation under oxidative stress is due to defects in autophagy. J. Cell Biochem. 2013;114:212–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

