Myeloid cell interferon responses correlate with clearance of SARS-CoV-2
- PMID: 35115549
- PMCID: PMC8814034
- DOI: 10.1038/s41467-022-28315-7
Myeloid cell interferon responses correlate with clearance of SARS-CoV-2
Abstract
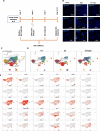
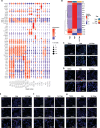
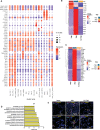
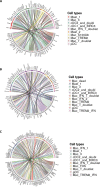
Emergence of mutant SARS-CoV-2 strains associated with an increased risk of COVID-19-related death necessitates better understanding of the early viral dynamics, host responses and immunopathology. Single cell RNAseq (scRNAseq) allows for the study of individual cells, uncovering heterogeneous and variable responses to environment, infection and inflammation. While studies have reported immune profiling using scRNAseq in terminal human COVID-19 patients, performing longitudinal immune cell dynamics in humans is challenging. Macaques are a suitable model of SARS-CoV-2 infection. Our longitudinal scRNAseq of bronchoalveolar lavage (BAL) cell suspensions from young rhesus macaques infected with SARS-CoV-2 (n = 6) demonstrates dynamic changes in transcriptional landscape 3 days post- SARS-CoV-2-infection (3dpi; peak viremia), relative to 14-17dpi (recovery phase) and pre-infection (baseline) showing accumulation of distinct populations of both macrophages and T-lymphocytes expressing strong interferon-driven inflammatory gene signature at 3dpi. Type I interferon response is induced in the plasmacytoid dendritic cells with appearance of a distinct HLADR+CD68+CD163+SIGLEC1+ macrophage population exhibiting higher angiotensin-converting enzyme 2 (ACE2) expression. These macrophages are significantly enriched in the lungs of macaques at 3dpi and harbor SARS-CoV-2 while expressing a strong interferon-driven innate anti-viral gene signature. The accumulation of these responses correlated with decline in viremia and recovery.
© 2022. The Author(s).
Conflict of interest statement
The authors declare competing interests.
Figures



Update of
-
Myeloid cell interferon responses correlate with clearance of SARS-CoV-2.Res Sq [Preprint]. 2021 Jul 15:rs.3.rs-664507. doi: 10.21203/rs.3.rs-664507/v1. Res Sq. 2021. Update in: Nat Commun. 2022 Feb 3;13(1):679. doi: 10.1038/s41467-022-28315-7. PMID: 34282414 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

