Answer ALS, a large-scale resource for sporadic and familial ALS combining clinical and multi-omics data from induced pluripotent cell lines
- PMID: 35115730
- PMCID: PMC8825283
- DOI: 10.1038/s41593-021-01006-0
Answer ALS, a large-scale resource for sporadic and familial ALS combining clinical and multi-omics data from induced pluripotent cell lines
Abstract
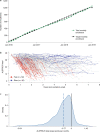
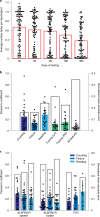
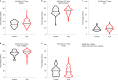
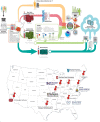
Answer ALS is a biological and clinical resource of patient-derived, induced pluripotent stem (iPS) cell lines, multi-omic data derived from iPS neurons and longitudinal clinical and smartphone data from over 1,000 patients with ALS. This resource provides population-level biological and clinical data that may be employed to identify clinical-molecular-biochemical subtypes of amyotrophic lateral sclerosis (ALS). A unique smartphone-based system was employed to collect deep clinical data, including fine motor activity, speech, breathing and linguistics/cognition. The iPS spinal neurons were blood derived from each patient and these cells underwent multi-omic analytics including whole-genome sequencing, RNA transcriptomics, ATAC-sequencing and proteomics. The intent of these data is for the generation of integrated clinical and biological signatures using bioinformatics, statistics and computational biology to establish patterns that may lead to a better understanding of the underlying mechanisms of disease, including subgroup identification. A web portal for open-source sharing of all data was developed for widespread community-based data analytics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. R.N., C.A. and G.A.C. disclose that their employer, IBM Research, is the research branch of IBM Corporation. R.N, C.A. and G.A.C. own stock in IBM Corporation.
Figures




References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

