Optimization of internal reference genes for qPCR in human pancreatic cancer research
- PMID: 35117652
- PMCID: PMC8797320
- DOI: 10.21037/tcr.2020.02.48
Optimization of internal reference genes for qPCR in human pancreatic cancer research
Abstract
Background: Pancreatic cancer (PC) has been becoming a common cancer with high mortality and quantitative real-time polymerase chain reaction (qPCR) is one of the best choices for researching gene expression. Internal reference genes, such as actin beta (ACTB) and glyceraldehyde-3-phosphatide hydrogenase (GAPDH) have long been used in relative quantification analysis. But evidence shows that some internal reference genes expression may vary in different tissues, cell lines and different conditions. The present study aimed to find more stable internal reference gene for qPCR experiment in PC.
Methods: Total RNA of human PC tissues were prepared using TRIZOL reagent. qPCR was performed using FastStart Universal SYBR Green Master to reflects the expression of target genes. Normfinder and geNorm were used to analyze the stability of chosen internal reference genes.
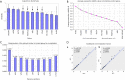
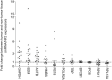
Results: According to the results of NormFinder and geNorm, eukaryotic translation initiation factor 2B subunit alpha (EIF2B1) and importin 8 (IPO8) were the same most stable internal reference genes in PCs and non-neoplastic tissues. In addition, EIF2B1 and IPO8 remained the most stable internal reference genes only in PCs. Using a normalization factor NF2 by geNorm as reference, the normalized GAPDH and ACTB expression levels were obviously up-regulated by 3.29- and 2.23-fold change, meanwhile ribosomal protein S17 (RPS17) were down-regulated by 0.77-fold change in PCs comparing with corresponding adjacent tissues.
Conclusions: The use of the combination of EIF2B1 and IPO8 would provide more stable results in differential expression analysis and prognostic analysis of PC.
Keywords: Pancreatic cancer (PC); internal reference gene; normalization; quantitative real-time polymerase chain reaction (qPCR).
2020 Translational Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.48). The authors have no conflicts of interest to declare.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
