Single-Cell Monitoring of Activated Innate Immune Signaling by a d2eGFP-Based Reporter Mimicking Time-Restricted Activation of IFNB1 Expression
- PMID: 35118008
- PMCID: PMC8803904
- DOI: 10.3389/fcimb.2021.784762
Single-Cell Monitoring of Activated Innate Immune Signaling by a d2eGFP-Based Reporter Mimicking Time-Restricted Activation of IFNB1 Expression
Abstract
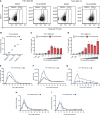
The innate immune system represents a balanced first line of defense against infection. Type I interferons (IFNs) are key regulators of the response to viral infections with an essential early wave of IFN-β expression, which is conditional, time-restricted, and stochastic in its nature. The possibility to precisely monitor individual cells with active IFNB1 transcription during innate signaling requires a robust reporter system that mimics the endogenous IFN-β signal. Here, we present a reporter system based on expression of a destabilized version of eGFP (d2eGFP) from a stably integrated reporter cassette containing the IFNB1 promoter and 3'-untranslated region, enabling both spatial and temporal detection of regulated IFNB1 expression. Specifically, this reporter permits detection, quantification, and isolation of cells actively producing d2eGFP in a manner that fully mimics IFN-β production allowing tracking of IFNB1 gene activation and repression in monocytic cells and keratinocytes. Using induced d2eGFP expression as a readout for activated immune signaling at the single-cell level, we demonstrate the application of the reporter for FACS-based selection of cells with genotypes supporting cGAS-STING signaling. Our studies provide a novel approach for monitoring on/off-switching of innate immune signaling and form the basis for investigating genotypes affecting immune regulation at the single-cell level.
Keywords: IFNB1 reporter; IFNB1 transcription; flow cytometry; innate immunity; single-cell.
Copyright © 2022 Thomsen, Andersen, Marqvorsen, Skipper, Paludan and Mikkelsen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


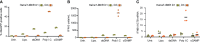

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

