Antibody variable sequences have a pronounced effect on cellular transport and plasma half-life
- PMID: 35118359
- PMCID: PMC8800109
- DOI: 10.1016/j.isci.2022.103746
Antibody variable sequences have a pronounced effect on cellular transport and plasma half-life
Abstract
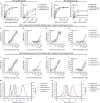
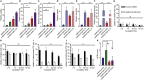
Monoclonal IgG antibodies are the fastest growing class of biologics, but large differences exist in their plasma half-life in humans. Thus, to design IgG antibodies with favorable pharmacokinetics, it is crucial to identify the determinants of such differences. Here, we demonstrate that the variable region sequences of IgG antibodies greatly affect cellular uptake and subsequent recycling and rescue from intracellular degradation by endothelial cells. When the variable sequences are masked by the cognate antigen, it influences both their transport behavior and binding to the neonatal Fc receptor (FcRn), a key regulator of IgG plasma half-life. Furthermore, we show how charge patch differences in the variable domains modulate both binding and transport properties and that a short plasma half-life, due to unfavorable charge patches, may partly be overcome by Fc-engineering for improved FcRn binding.
Keywords: Biological sciences; Biophysics; Immunology.
© 2022 The Author(s).
Conflict of interest statement
T.S., T.E., F.A.A.J., and A.G. are under paid employment by the company F. Hoffmann-La Roche. Other authors declare no conflict of interest.
Figures




References
-
- Andersen J.T., Justesen S., Fleckenstein B., Michaelsen T.E., Berntzen G., Kenanova V.E., Daba M.B., Lauvrak V., Buus S., Sandlie I. Ligand binding and antigenic properties of a human neonatal Fc receptor with mutation of two unpaired cysteine residues. FEBS J. 2008;275:4097–4110. - PubMed
-
- Bloch Y., Bouchareychas L., Merceron R., Adanowska K., Bossche L., Detry S., Govindarajan S., Elewaut D., Haerynck F., Dullaers M. Structural activation of pro-inflammatory human cytokine IL-23 by cognate IL-23 receptor enables recruitment of the shared receptor IL-12r 1. Immunity. 2017;48:45–58. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

