The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant
- PMID: 35118687
- PMCID: PMC9015498
- DOI: 10.1002/jmv.27643
The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant
Abstract
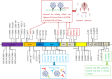
Recently, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant (B.1.1.529) was first identified in Botswana in November 2021. It was first reported to the World Health Organization (WHO) on November 24. On November 26, 2021, according to the advice of scientists who are part of the WHO's Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAG-VE), the WHO defined the strain as a variant of concern (VOC) and named it Omicron. Compared to the other four VOCs (Alpha, Beta, Gamma, and Delta), the Omicron variant was the most highly mutated strain, with 50 mutations accumulated throughout the genome. The Omicron variant contains at least 32 mutations in the spike protein, which was twice as many as the Delta variant. Studies have shown that carrying many mutations can increase infectivity and immune escape of the Omicron variant compared with the early wild-type strain and the other four VOCs. The Omicron variant is becoming the dominant strain in many countries worldwide and brings new challenges to preventing and controlling coronavirus disease 2019 (COVID-19). The current review article aims to analyze and summarize information data about the biological characteristics of amino acid mutations, the epidemic characteristics, immune escape, and vaccine reactivity of the Omicron variant, hoping to provide a scientific reference for monitoring, prevention, and vaccine development strategies for the Omicron variant.
Keywords: B.1.1.529; COVID-19; Omicron; SARS-CoV-2; immune escape.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

