Assembly-free rapid differential gene expression analysis in non-model organisms using DNA-protein alignment
- PMID: 35120462
- PMCID: PMC8815227
- DOI: 10.1186/s12864-021-08278-7
Assembly-free rapid differential gene expression analysis in non-model organisms using DNA-protein alignment
Abstract
Background: RNA-seq is being increasingly adopted for gene expression studies in a panoply of non-model organisms, with applications spanning the fields of agriculture, aquaculture, ecology, and environment. For organisms that lack a well-annotated reference genome or transcriptome, a conventional RNA-seq data analysis workflow requires constructing a de-novo transcriptome assembly and annotating it against a high-confidence protein database. The assembly serves as a reference for read mapping, and the annotation is necessary for functional analysis of genes found to be differentially expressed. However, assembly is computationally expensive. It is also prone to errors that impact expression analysis, especially since sequencing depth is typically much lower for expression studies than for transcript discovery.
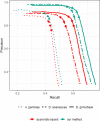
Results: We propose a shortcut, in which we obtain counts for differential expression analysis by directly aligning RNA-seq reads to the high-confidence proteome that would have been otherwise used for annotation. By avoiding assembly, we drastically cut down computational costs - the running time on a typical dataset improves from the order of tens of hours to under half an hour, and the memory requirement is reduced from the order of tens of Gbytes to tens of Mbytes. We show through experiments on simulated and real data that our pipeline not only reduces computational costs, but has higher sensitivity and precision than a typical assembly-based pipeline. A Snakemake implementation of our workflow is available at: https://bitbucket.org/project_samar/samar .
Conclusions: The flip side of RNA-seq becoming accessible to even modestly resourced labs has been that the time, labor, and infrastructure cost of bioinformatics analysis has become a bottleneck. Assembly is one such resource-hungry process, and we show here that it can be avoided for quick and easy, yet more sensitive and precise, differential gene expression analysis in non-model organisms.
Keywords: DNA-protein alignment; Differential gene expression analysis; Non-model organisms; RNA-seq; Transcriptome assembly.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Stark R, Grzelak M, Hadfield J. Rna sequencing: the teenage years. Nat Rev Genet. 2019;20:631–56. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

