Oligomerization of DNA replication regulatory protein RADX is essential to maintain replication fork stability
- PMID: 35120927
- PMCID: PMC8902620
- DOI: 10.1016/j.jbc.2022.101672
Oligomerization of DNA replication regulatory protein RADX is essential to maintain replication fork stability
Abstract
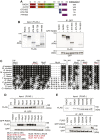
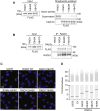
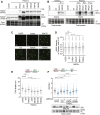
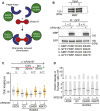
Genome integrity requires complete and accurate DNA replication once per cell division cycle. Replication stress poses obstacles to this process that must be overcome to prevent replication fork collapse. An important regulator of replication fork stability is the RAD51 protein, which promotes replication fork reversal and protects nascent DNA strands from nuclease-mediated degradation. Many regulatory proteins control these RAD51 activities, including RADX, which binds both ssDNA and RAD51 at replication forks to ensure that fork reversal is confined to stalled forks. Many ssDNA-binding proteins function as hetero- or homo-oligomers. In this study, we addressed whether this is also the case for RADX. Using biochemical and genetic approaches, we found that RADX acts as a homo-oligomer to control replication fork stability. RADX oligomerizes using at least two different interaction surfaces, including one mapped to a C-terminal region. We demonstrate that mutations in this region prevent oligomerization and prevent RADX function in cells, and that addition of a heterologous dimerization domain to the oligomerization mutants restored their ability to regulate replication. Taken together, our results demonstrate that like many ssDNA-binding proteins, oligomerization is essential for RADX-mediated regulation of genome stability.
Keywords: DNA combing; DNA repair; RAD51; RADX; RPA; cxorf57; dimerization; fork protection; fork reversal; genome stability.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interests with the contents of this article.
Figures

References
-
- Berti M., Cortez D., Lopes M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 2020;21:633–651. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

