Fluoxetine ameliorates mucopolysaccharidosis type IIIA
- PMID: 35121108
- PMCID: PMC9077373
- DOI: 10.1016/j.ymthe.2022.01.037
Fluoxetine ameliorates mucopolysaccharidosis type IIIA
Abstract
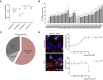
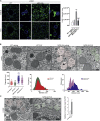
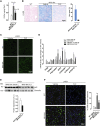
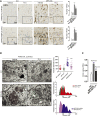
Mucopolysaccharidosis type IIIA (MPS-IIIA) is an autosomal recessive disorder caused by mutations in SGSH involved in the degradation of heparan sulfate. MPS-IIIA presents severe neurological symptoms such as progressive developmental delay and cognitive decline, for which there is currently no treatment. Brain targeting represents the main challenge for therapeutics to treat MPS-IIIA, and the development of small-molecule-based treatments able to reach the CNS could be a relevant advance for therapy. Using cell-based high content imaging to survey clinically approved drugs in MPS-IIIA cells, we identified fluoxetine, a selective serotonin reuptake inhibitor. Fluoxetine increases lysosomal and autophagic functions via TFEB activation through a RagC-dependent mechanism. Mechanistically, fluoxetine increases lysosomal exocytosis in mouse embryonic fibroblasts from MPS-IIIA mice, suggesting that this process may be responsible for heparan sulfate clearance. In vivo, fluoxetine ameliorates somatic and brain pathology in a mouse model of MPS-IIIA by decreasing the accumulation of glycosaminoglycans and aggregated autophagic substrates, reducing inflammation, and slowing down cognitive deterioration. We repurposed fluoxetine for potential therapeutics to treat human MPS-IIIA disease.
Keywords: MPS-IIIA; TFEB; autophagy; drug repurposing; fluoxetine; high content imaging; lysosomal exocytosis; lysosomal storage disorders.
Copyright © 2022 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Monaco A., Maffia V., Sorrentino N.C., Sambri I., Ezhova Y., Giuliano T., Cacace V., Nusco E., De Risi M., De Leonibus E., et al. The amyloid inhibitor CLR01 relieves autophagy and ameliorates neuropathology in a severe lysosomal storage disease. Mol. Ther. 2020;28:1167–1176. doi: 10.1016/j.ymthe.2020.02.005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

