Ionic mitigation of CD4+ T cell metabolic fitness, Th1 central nervous system autoimmunity and Th2 asthmatic airway inflammation by therapeutic zinc
- PMID: 35121767
- PMCID: PMC8816938
- DOI: 10.1038/s41598-022-04827-6
Ionic mitigation of CD4+ T cell metabolic fitness, Th1 central nervous system autoimmunity and Th2 asthmatic airway inflammation by therapeutic zinc
Abstract
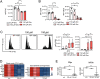
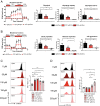
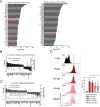
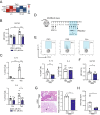
T helper (Th) cells provide immunity to pathogens but also contribute to detrimental immune responses during allergy and autoimmunity. Th2 cells mediate asthmatic airway inflammation and Th1 cells are involved in the pathogenesis of multiple sclerosis. T cell activation involves complex transcriptional networks and metabolic reprogramming, which enable proliferation and differentiation into Th1 and Th2 cells. The essential trace element zinc has reported immunomodulatory capacity and high zinc concentrations interfere with T cell function. However, how high doses of zinc affect T cell gene networks and metabolism remained so far elusive. Herein, we demonstrate by means of transcriptomic analysis that zinc aspartate (UNIZINK), a registered pharmaceutical infusion solution with high bioavailability, negatively regulates gene networks controlling DNA replication and the energy metabolism of murine CD3/CD28-activated CD4+ T cells. Specifically, in the presence of zinc, CD4+ T cells show impaired expression of cell cycle, glycolytic and tricarboxylic acid cycle genes, which functionally cumulates in reduced glycolysis, oxidative phosphorylation, metabolic fitness and viability. Moreover, high zinc concentrations impaired nuclear expression of the metabolic transcription factor MYC, prevented Th1 and Th2 differentiation in vitro and reduced Th1 autoimmune central nervous system (CNS) inflammation and Th2 asthmatic airway inflammation induced by house dust mites in vivo. Together, we find that higher zinc doses impair the metabolic fitness of CD4+ T cells and prevent Th1 CNS autoimmunity and Th2 allergy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Ruterbusch M, et al. In vivo CD4(+) T cell differentiation and function: Revisiting the Th1/Th2 paradigm. Annu. Rev. Immunol. 2020;38:705–725. - PubMed
-
- Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. 2021;184(9):2521–2522. - PubMed
-
- Mattiuzzi C, Lippi G. Worldwide asthma epidemiology: insights from the global health data exchange database. Int. Forum Allergy Rhinol. 2020;10(1):75–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

