RNF2 ablation reprograms the tumor-immune microenvironment and stimulates durable NK and CD4+ T-cell-dependent antitumor immunity
- PMID: 35121884
- PMCID: PMC8809507
- DOI: 10.1038/s43018-021-00263-z
RNF2 ablation reprograms the tumor-immune microenvironment and stimulates durable NK and CD4+ T-cell-dependent antitumor immunity
Abstract
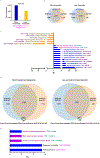
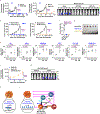
Expanding the utility of immune-based cancer treatments is a clinical challenge due to tumor-intrinsic factors that suppress the immune response. Here we report the identification of tumoral ring finger protein 2 (RNF2), the core subunit of polycomb repressor complex 1, as a negative regulator of antitumor immunity in various human cancers, including breast cancer. In syngeneic murine models of triple-negative breast cancer, we found that deleting genes encoding the polycomb repressor complex 1 subunits Rnf2, BMI1 proto-oncogene, polycomb ring finger (Bmi1), or the downstream effector of Rnf2, remodeling and spacing factor 1 (Rsf1), was sufficient by itself to induce durable tumor rejection and establish immune memory by enhancing infiltration and activation of natural killer and CD4+ T cells, but not CD8+ T cells, into the tumor and enabled their cooperativity. These findings uncover an epigenetic reprogramming of the tumor-immune microenvironment, which fosters durable antitumor immunity and memory.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures









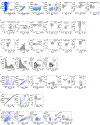






Comment in
-
A PRC1-RNF2 knockout punch for cancer.Nat Cancer. 2021 Oct;2(10):996-997. doi: 10.1038/s43018-021-00270-0. Nat Cancer. 2021. PMID: 35121885 No abstract available.
References
Methods-only references
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

